Species boundaries in the human pathogen Paracoccidioides
- PMID: 28602831
- PMCID: PMC8335726
- DOI: 10.1016/j.fgb.2017.05.007
Species boundaries in the human pathogen Paracoccidioides
Abstract
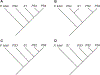
The use of molecular taxonomy for identifying recently diverged species has transformed the study of speciation in fungi. The pathogenic fungus Paracoccidioides spp has been hypothesized to be composed of five phylogenetic species, four of which compose the brasiliensis species complex. Nuclear gene genealogies support this divergence scenario, but mitochondrial loci do not; while all species from the brasiliensis complex are differentiated at nuclear coding loci, they are not at mitochondrial loci. We addressed the source of this incongruity using 11 previously published gene fragments, 10 newly-sequenced nuclear non-coding loci, and 10 microsatellites. We hypothesized and further demonstrated that the mito-nuclear incongruence in the brasiliensis species complex results from interspecific hybridization and mitochondrial introgression, a common phenomenon in eukaryotes. Additional population genetic analyses revealed possible nuclear introgression but much less than that seen in the mitochondrion. Our results are consistent with a divergence scenario of secondary contact and subsequent mitochondrial introgression despite the continued persistence of species boundaries. We also suggest that yeast morphology slightly-but significantly-differs across all five Paracoccidioides species and propose to elevate four of these phylogenetic species to formally described taxonomic species.
Keywords: Introgression; Paracoccidioides spp; Speciation.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
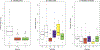
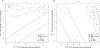

References
-
- Abbott R, Albach D, Ansell S, et al. , 2013. Hybridization and speciation. J. Evol. Biol 26, 229–246. - PubMed
-
- Ajello L, Cheng S-L, 1967. Sexual reproduction in Histoplasma capsulatum. Mycologia 59, 689. - PubMed
-
- Akaike H, 1974. A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723.
-
- Almeida AJ, Cunha C, Carmona JA, et al. , 2009. Cdc42p controls yeast-cell shape and virulence of Paracoccidioides brasiliensis. Fungal Genet. Biol 46, 919–926. - PubMed
-
- Almeida AJ, Matute DR, Carmona JA, et al. , 2007. Genome size and ploidy of Paracoccidioides brasiliensis reveals a haploid DNA content: flow cytometry and GP43 sequence analysis. Fungal Genet. Biol 44, 25–31. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

