Structural Characterization of Maize SIRK1 Kinase Domain Reveals an Unusual Architecture of the Activation Segment
- PMID: 28603531
- PMCID: PMC5445127
- DOI: 10.3389/fpls.2017.00852
Structural Characterization of Maize SIRK1 Kinase Domain Reveals an Unusual Architecture of the Activation Segment
Abstract
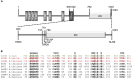
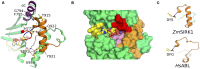
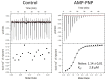
Kinases are primary regulators of plant metabolism and excellent targets for plant breeding. However, most kinases, including the abundant receptor-like kinases (RLK), have no assigned role. SIRK1 is a leucine-rich repeat receptor-like kinase (LRR-RLK), the largest family of RLK. In Arabidopsis thaliana, SIRK1 (AtSIRK1) is phosphorylated after sucrose is resupplied to sucrose-starved seedlings and it modulates the sugar response by phosphorylating several substrates. In maize, the ZmSIRK1 expression is altered in response to drought stress. In neither Arabidopsis nor in maize has the function of SIRK1 been completely elucidated. As a first step toward the biochemical characterization of ZmSIRK1, we obtained its recombinant kinase domain, demonstrated that it binds AMP-PNP, a non-hydrolysable ATP-analog, and solved the structure of ZmSIRK1- AMP-PNP co-crystal. The ZmSIRK1 crystal structure revealed a unique conformation for the activation segment. In an attempt to find inhibitors for ZmSIRK1, we screened a focused small molecule library and identified six compounds that stabilized ZmSIRK1 against thermal melt. ITC analysis confirmed that three of these compounds bound to ZmSIRK1 with low micromolar affinity. Solving the 3D structure of ZmSIRK1-AMP-PNP co-crystal provided information on the molecular mechanism of ZmSIRK1 activity. Furthermore, the identification of small molecules that bind this kinase can serve as initial backbone for development of new potent and selective ZmSIRK1 antagonists.
Keywords: SIRK1; ligand; maize; receptor-like kinase; structure.
Figures



References
-
- Albert M., Jehle A. K., Mueller K., Eisele C., Lipschis M., Felix G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285 19035–19042. 10.1074/jbc.M110.124800 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

