Distinct DNA methylation profiles in subtypes of orofacial cleft
- PMID: 28603561
- PMCID: PMC5465456
- DOI: 10.1186/s13148-017-0362-2
Distinct DNA methylation profiles in subtypes of orofacial cleft
Abstract
Background: Epigenetic data could help identify risk factors for orofacial clefts, either by revealing a causal role for epigenetic mechanisms in causing clefts or by capturing information about causal genetic or environmental factors. Given the evidence that different subtypes of orofacial cleft have distinct aetiologies, we explored whether children with different cleft subtypes showed distinct epigenetic profiles.
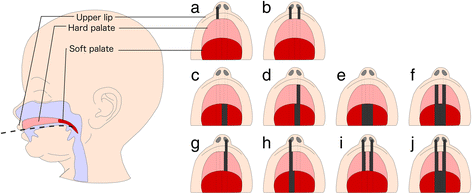
Methods: In whole-blood samples from 150 children from the Cleft Collective cohort study, we measured DNA methylation at over 450,000 sites on the genome. We then carried out epigenome-wide association studies (EWAS) to test the association between methylation at each site and cleft subtype (cleft lip only (CLO) n = 50; cleft palate only (CPO) n = 50; cleft lip and palate (CLP) n = 50). We also compared methylation in the blood to methylation in the lip or palate tissue using genome-wide data from the same 150 children and conducted an EWAS of CLO compared to CLP in lip tissue.
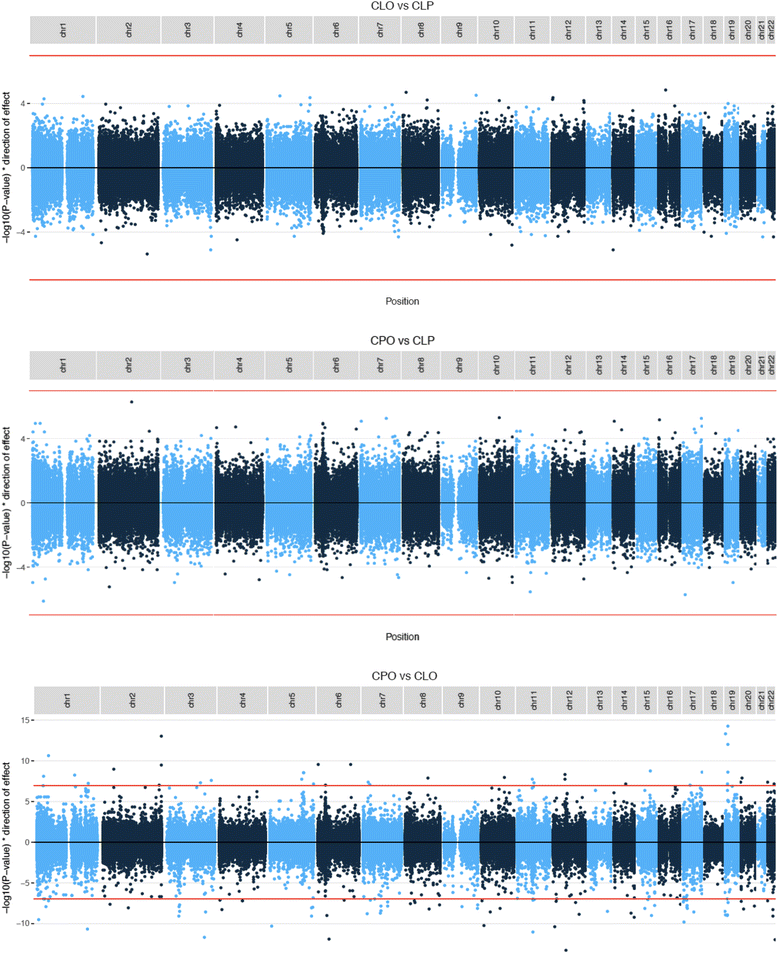
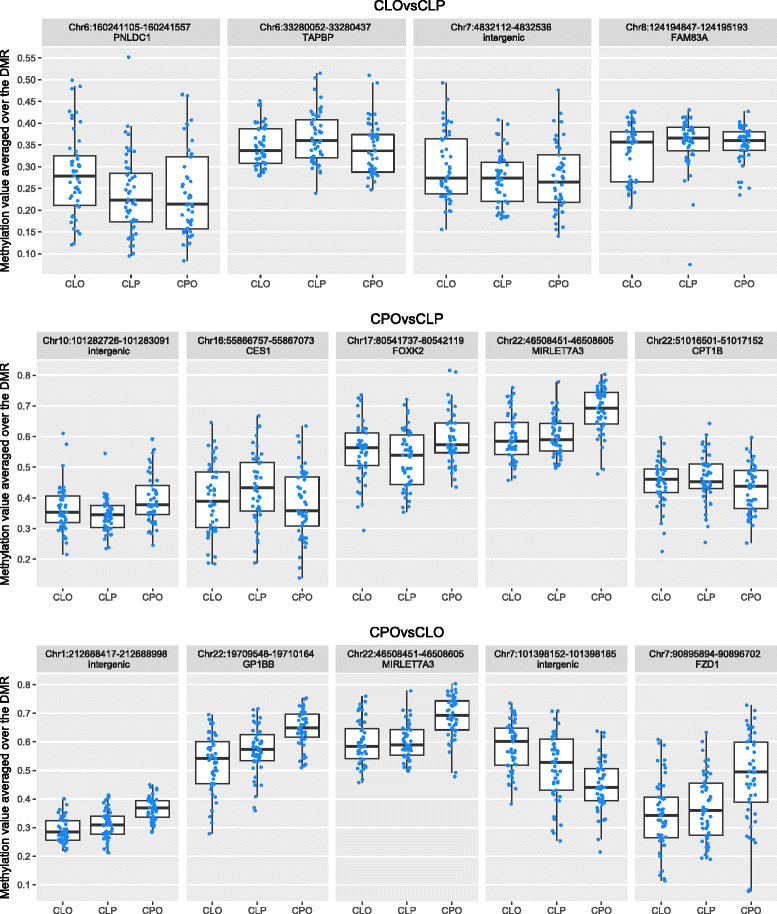
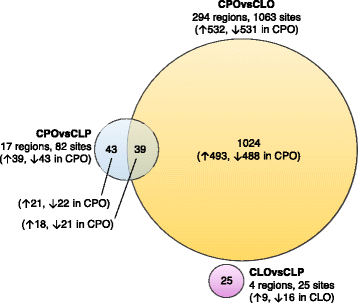
Results: We found four genomic regions in blood differentially methylated in CLO compared to CLP, 17 in CPO compared to CLP and 294 in CPO compared to CLO. Several regions mapped to genes that have previously been implicated in the development of orofacial clefts (for example, TBX1, COL11A2, HOXA2, PDGFRA), and over 250 associations were novel. Methylation in blood correlated with that in lip/palate at some regions. There were 14 regions differentially methylated in the lip tissue from children with CLO and CLP, with one region (near KIAA0415) showing up in both the blood and lip EWAS.
Conclusions: Our finding of distinct methylation profiles in different orofacial cleft (OFC) subtypes represents a promising first step in exploring the potential role of epigenetic modifications in the aetiology of OFCs and/or as clinically useful biomarkers of OFC subtypes.
Keywords: Cleft Collective; Cleft lip; Cleft palate; DNA methylation; EWAS; Epigenome-wide association study; Orofacial clefts.
Figures

References
-
- EUROCAT . EUROCAT Website Database. 2017.
-
- Sharp GC, Stergiakouli E, Sandy J, Relton C. Epigenetics and orofacial clefts: a brief introduction. Cleft Palate Craniofac J. 2017 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

