Defining Disease, Diagnosis, and Translational Medicine within a Homeostatic Perturbation Paradigm: The National Institutes of Health Undiagnosed Diseases Program Experience
- PMID: 28603714
- PMCID: PMC5445140
- DOI: 10.3389/fmed.2017.00062
Defining Disease, Diagnosis, and Translational Medicine within a Homeostatic Perturbation Paradigm: The National Institutes of Health Undiagnosed Diseases Program Experience
Abstract
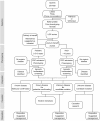
Traditionally, the use of genomic information for personalized medical decisions relies on prior discovery and validation of genotype-phenotype associations. This approach constrains care for patients presenting with undescribed problems. The National Institutes of Health (NIH) Undiagnosed Diseases Program (UDP) hypothesized that defining disease as maladaptation to an ecological niche allows delineation of a logical framework to diagnose and evaluate such patients. Herein, we present the philosophical bases, methodologies, and processes implemented by the NIH UDP. The NIH UDP incorporated use of the Human Phenotype Ontology, developed a genomic alignment strategy cognizant of parental genotypes, pursued agnostic biochemical analyses, implemented functional validation, and established virtual villages of global experts. This systematic approach provided a foundation for the diagnostic or non-diagnostic answers provided to patients and serves as a paradigm for scalable translational research.
Keywords: diploid alignment; distributed cognition; glycome; human phenotype ontology; rare disease.
Figures
References
-
- Caudle KE, Klein TE, Hoffman JM, Muller DJ, Whirl-Carrillo M, Gong L, et al. Incorporation of pharmacogenomics into routine clinical practice: the clinical pharmacogenetics implementation consortium (CPIC) guideline development process. Curr Drug Metab (2014) 15:209–17.10.2174/1389200215666140130124910 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

