Androgen Receptor-Dependent and -Independent Mechanisms Involved in Prostate Cancer Therapy Resistance
- PMID: 28604629
- PMCID: PMC5483886
- DOI: 10.3390/cancers9060067
Androgen Receptor-Dependent and -Independent Mechanisms Involved in Prostate Cancer Therapy Resistance
Abstract
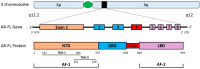
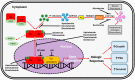
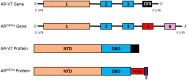
Despite the initial efficacy of androgen deprivation in prostate cancer, virtually all patients progress to castration-resistant prostate cancer (CRPC). Androgen receptor (AR) signaling is critically required for CRPC. A new generation of medications targeting AR, such as abiraterone and enzalutamide, has improved survival of metastatic CRPC (mCRPC) patients. However, a significant proportion of patients presents with primary resistance to these agents, and in the remainder, secondary resistance will invariably develop, which makes mCRPC the lethal form of the disease. Mechanisms underlying progression to mCRPC and treatment resistance are extremely complex. AR-dependent resistance mechanisms include AR amplification, AR point mutations, expression of constitutively active AR splice variants, and altered intratumoral androgen biosynthesis. AR-independent resistance mechanisms include glucocorticoid receptor activation, immune-mediated resistance, and neuroendocrine differentiation. The development of novel agents, such as seviteronel, apalutamide, and EPI-001/EPI-506, as well as the identification and validation of novel predictive biomarkers of resistance, may lead to improved therapeutics for mCRPC patients.
Keywords: abiraterone; androgen receptor; castration-resistant prostate cancer; enzalutamide; progression; resistance mechanisms.
Conflict of interest statement
Young E. Whang received research funding from Astellas, Janssen, and Tokai. Daniel J. Crona stated no conflicts of interest.
Figures

References
-
- American Cancer Society . Cancer Facts & Figures 2017. American Cancer Society; Atlanta, GA, USA: 2017.
-
- Howlader N., Noone A.M., Krapcho M., Miller D., Bishop K., Kosary C.L., Yu M., Ruhl J., Tatalovich Z., Mariotto A., et al. Seercancer Statistics Review, 1975–2012. National Cancer Institute; Bethesda, MD, USA: 2015.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

