Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor
- PMID: 28604728
- PMCID: PMC6016736
- DOI: 10.1038/ng.3896
Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor
Abstract
Genome-wide association studies (GWAS) have transformed understanding of susceptibility to testicular germ cell tumors (TGCTs), but much of the heritability remains unexplained. Here we report a new GWAS, a meta-analysis with previous GWAS and a replication series, totaling 7,319 TGCT cases and 23,082 controls. We identify 19 new TGCT risk loci, roughly doubling the number of known TGCT risk loci to 44. By performing in situ Hi-C in TGCT cells, we provide evidence for a network of physical interactions among all 44 TGCT risk SNPs and candidate causal genes. Our findings implicate widespread disruption of developmental transcriptional regulators as a basis of TGCT susceptibility, consistent with failed primordial germ cell differentiation as an initiating step in oncogenesis. Defective microtubule assembly and dysregulation of KIT-MAPK signaling also feature as recurrently disrupted pathways. Our findings support a polygenic model of risk and provide insight into the biological basis of TGCT.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
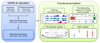
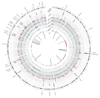
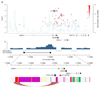


Comment in
-
Re: Identification of 19 New Risk Loci and Potential Regulatory Mechanisms Influencing Susceptibility to Testicular Germ Cell Tumor.J Urol. 2018 Oct;200(4):700. doi: 10.1016/j.juro.2018.06.047. Epub 2018 Jul 7. J Urol. 2018. PMID: 30227584 No abstract available.
References
-
- Manku G, et al. Changes in the expression profiles of claudins during gonocyte differentiation and in seminomas. Andrology. 2016;4:95–110. - PubMed
-
- Le Cornet C, et al. Testicular cancer incidence to rise by 25% by 2025 in Europe? Model-based predictions in 40 countries using population-based registry data. Eur J Cancer. 2014;50:831–9. - PubMed
-
- Swerdlow AJ, De Stavola BL, Swanwick MA, Maconochie NE. Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet. 1997;350:1723–8. - PubMed
-
- McGlynn KA, Devesa SS, Graubard BI, Castle PE. Increasing incidence of testicular germ cell tumors among black men in the United States. J Clin Oncol. 2005;23:5757–61. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- G1001799/MRC_/Medical Research Council/United Kingdom
- 17528/CRUK_/Cancer Research UK/United Kingdom
- MR/N01104X/2/MRC_/Medical Research Council/United Kingdom
- U01 CA188392/CA/NCI NIH HHS/United States
- G0700491/MRC_/Medical Research Council/United Kingdom
- U19 CA148537/CA/NCI NIH HHS/United States
- 15007/CRUK_/Cancer Research UK/United Kingdom
- 16563/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 10589/CRUK_/Cancer Research UK/United Kingdom
- 13065/CRUK_/Cancer Research UK/United Kingdom
- 19167/CRUK_/Cancer Research UK/United Kingdom
- MR/N01104X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

