The Genomic Impact of Gene Retrocopies: What Have We Learned from Comparative Genomics, Population Genomics, and Transcriptomic Analyses?
- PMID: 28605529
- PMCID: PMC5470649
- DOI: 10.1093/gbe/evx081
The Genomic Impact of Gene Retrocopies: What Have We Learned from Comparative Genomics, Population Genomics, and Transcriptomic Analyses?
Abstract
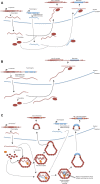
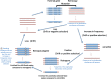
Gene duplication is a major driver of organismal evolution. Gene retroposition is a mechanism of gene duplication whereby a gene's transcript is used as a template to generate retroposed gene copies, or retrocopies. Intriguingly, the formation of retrocopies depends upon the enzymatic machinery encoded by retrotransposable elements, genomic parasites occurring in the majority of eukaryotes. Most retrocopies are depleted of the regulatory regions found upstream of their parental genes; therefore, they were initially considered transcriptionally incompetent gene copies, or retropseudogenes. However, examples of functional retrocopies, or retrogenes, have accumulated since the 1980s. Here, we review what we have learned about retrocopies in animals, plants and other eukaryotic organisms, with a particular emphasis on comparative and population genomic analyses complemented with transcriptomic datasets. In addition, these data have provided information about the dynamics of the different "life cycle" stages of retrocopies (i.e., polymorphic retrocopy number variants, fixed retropseudogenes and retrogenes) and have provided key insights into the retroduplication mechanisms, the patterns and evolutionary forces at work during the fixation process and the biological function of retrogenes. Functional genomic and transcriptomic data have also revealed that many retropseudogenes are transcriptionally active and a biological role has been experimentally determined for many. Finally, we have learned that not only non-long terminal repeat retroelements but also long terminal repeat retroelements play a role in the emergence of retrocopies across eukaryotes. This body of work has shown that mRNA-mediated duplication represents a widespread phenomenon that produces an array of new genes that contribute to organismal diversity and adaptation.
Keywords: new functions; pollen expression; regulatory element; retroCNV; retrocopy; retrogene; retropseudogene; testis expression.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Genomic exploration of retrocopies in Insect pests of plants and their role in the expansion of heat shock proteins superfamily as evolutionary targets.BMC Genomics. 2024 Nov 20;25(1):1116. doi: 10.1186/s12864-024-11056-w. BMC Genomics. 2024. PMID: 39567882 Free PMC article.
-
The life history of retrocopies illuminates the evolution of new mammalian genes.Genome Res. 2016 Mar;26(3):301-14. doi: 10.1101/gr.198473.115. Epub 2016 Jan 4. Genome Res. 2016. PMID: 26728716 Free PMC article.
-
A Genome-Wide Landscape of Retrocopies in Primate Genomes.Genome Biol Evol. 2015 Jul 29;7(8):2265-75. doi: 10.1093/gbe/evv142. Genome Biol Evol. 2015. PMID: 26224704 Free PMC article.
-
Protein-Coding Genes' Retrocopies and Their Functions.Viruses. 2017 Apr 13;9(4):80. doi: 10.3390/v9040080. Viruses. 2017. PMID: 28406439 Free PMC article. Review.
-
Not So Dead Genes-Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts.Cells. 2021 Apr 15;10(4):912. doi: 10.3390/cells10040912. Cells. 2021. PMID: 33921034 Free PMC article. Review.
Cited by
-
Structural characterization and duplication modes of pseudogenes in plants.Sci Rep. 2021 Mar 5;11(1):5292. doi: 10.1038/s41598-021-84778-6. Sci Rep. 2021. PMID: 33674668 Free PMC article.
-
Retrotransposition facilitated the establishment of a primary plastid in the thecate amoeba Paulinella.Proc Natl Acad Sci U S A. 2022 Jun 7;119(23):e2121241119. doi: 10.1073/pnas.2121241119. Epub 2022 May 31. Proc Natl Acad Sci U S A. 2022. PMID: 35639693 Free PMC article.
-
Retrocopying expands the functional repertoire of APOBEC3 antiviral proteins in primates.Elife. 2020 Jun 1;9:e58436. doi: 10.7554/eLife.58436. Elife. 2020. PMID: 32479260 Free PMC article.
-
Multiple FGF4 Retrocopies Recently Derived within Canids.Genes (Basel). 2020 Jul 23;11(8):839. doi: 10.3390/genes11080839. Genes (Basel). 2020. PMID: 32717834 Free PMC article.
-
Completion of LINE integration involves an open '4-way' branched DNA intermediate.Nucleic Acids Res. 2019 Sep 19;47(16):8708-8719. doi: 10.1093/nar/gkz673. Nucleic Acids Res. 2019. PMID: 31392993 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

