Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice
- PMID: 28605576
- PMCID: PMC5785341
- DOI: 10.1111/pbi.12771
Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice
Abstract
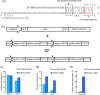
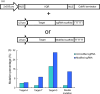
Clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) is a revolutionary technology that enables efficient genomic modification in many organisms. Currently, the wide use of Streptococcus pyogenes Cas9 (SpCas9) primarily recognizes sites harbouring a canonical NGG protospacer adjacent motif (PAM). The newly developed VQR (D1135V/R1335Q/T1337R) variant of Cas9 has been shown to cleave sites containing NGA PAM in rice, which greatly expanded the range of genome editing. However, the low editing efficiency of the VQR variant remains, which limits its wide application in genome editing. In this study, by modifying the single guide RNA (sgRNA) structure and strong endogenous promoters, we significantly increased the editing efficiency of the VQR variant. The modified CRISPR-Cas9-VQR system provides a robust toolbox for multiplex genome editing at sites containing noncanonical NGA PAM.
Keywords: CRISPR-Cas9; VQR; efficiency; genome editing; rice.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

Similar articles
-
[Cas9 protein variant VQR recognizes NGAC protospacer adjacent motif in rice].Yi Chuan. 2018 Dec 20;40(12):1112-1119. doi: 10.16288/j.yczz.18-126. Yi Chuan. 2018. PMID: 30559100 Chinese.
-
Cas9-NG Greatly Expands the Targeting Scope of the Genome-Editing Toolkit by Recognizing NG and Other Atypical PAMs in Rice.Mol Plant. 2019 Jul 1;12(7):1015-1026. doi: 10.1016/j.molp.2019.03.010. Epub 2019 Mar 27. Mol Plant. 2019. PMID: 30928635
-
Improving Plant Genome Editing with High-Fidelity xCas9 and Non-canonical PAM-Targeting Cas9-NG.Mol Plant. 2019 Jul 1;12(7):1027-1036. doi: 10.1016/j.molp.2019.03.011. Epub 2019 Mar 27. Mol Plant. 2019. PMID: 30928637
-
Cas9, Cpf1 and C2c1/2/3-What's next?Bioengineered. 2017 May 4;8(3):265-273. doi: 10.1080/21655979.2017.1282018. Epub 2017 Jan 31. Bioengineered. 2017. PMID: 28140746 Free PMC article. Review.
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
Cited by
-
Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice.BMC Plant Biol. 2019 Nov 21;19(1):511. doi: 10.1186/s12870-019-2131-1. BMC Plant Biol. 2019. PMID: 31752697 Free PMC article.
-
Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering.BMC Plant Biol. 2020 May 25;20(1):234. doi: 10.1186/s12870-020-02385-5. BMC Plant Biol. 2020. PMID: 32450802 Free PMC article. Review.
-
Unleashing the Potential of CRISPR/Cas9 Genome Editing for Yield-Related Traits in Rice.Plants (Basel). 2024 Oct 24;13(21):2972. doi: 10.3390/plants13212972. Plants (Basel). 2024. PMID: 39519891 Free PMC article. Review.
-
Enhanced FnCas12a-Mediated Targeted Mutagenesis Using crRNA With Altered Target Length in Rice.Front Genome Ed. 2020 Dec 14;2:608563. doi: 10.3389/fgeed.2020.608563. eCollection 2020. Front Genome Ed. 2020. PMID: 34713233 Free PMC article.
-
Engineering Abiotic Stress Tolerance in Crop Plants through CRISPR Genome Editing.Cells. 2022 Nov 13;11(22):3590. doi: 10.3390/cells11223590. Cells. 2022. PMID: 36429019 Free PMC article. Review.
References
-
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

