A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production
- PMID: 28605577
- PMCID: PMC5785352
- DOI: 10.1111/pbi.12772
A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production
Abstract
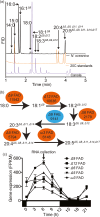
Nannochloropsis oceanica is an oleaginous microalga rich in ω3 long-chain polyunsaturated fatty acids (LC-PUFAs) content, in the form of eicosapentaenoic acid (EPA). We identified the enzymes involved in LC-PUFA biosynthesis in N. oceanica CCMP1779 and generated multigene expression vectors aiming at increasing LC-PUFA content in vivo. We isolated the cDNAs encoding four fatty acid desaturases (FAD) and determined their function by heterologous expression in S. cerevisiae. To increase the expression of multiple fatty acid desaturases in N. oceanica CCMP1779, we developed a genetic engineering toolkit that includes an endogenous bidirectional promoter and optimized peptide bond skipping 2A peptides. The toolkit also includes multiple epitopes for tagged fusion protein production and two antibiotic resistance genes. We applied this toolkit, towards building a gene stacking system for N. oceanica that consists of two vector series, pNOC-OX and pNOC-stacked. These tools for genetic engineering were employed to test the effects of the overproduction of one, two or three desaturase-encoding cDNAs in N. oceanica CCMP1779 and prove the feasibility of gene stacking in this genetically tractable oleaginous microalga. All FAD overexpressing lines had considerable increases in the proportion of LC-PUFAs, with the overexpression of Δ12 and Δ5 FAD encoding sequences leading to an increase in the final ω3 product, EPA.
Keywords: Nannochloropsis; 2A peptides; LC-PUFA; bidirectional promoters; eicosapentaenoic acid; gene stacking; genetic engineering toolkit; multigene expression.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures





References
-
- Broun, P. (1998) Catalytic plasticity of fatty acid modification enzymes underlying chemical diversity of plant lipids. Science, 282, 1315–1317. - PubMed
-
- Cahoon, E.B. , Shockey, J.M. , Dietrich, C.R. , Gidda, S.K. , Mullen, R.T. and Dyer, J.M. (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol. 10, 236–244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

