DNA polymerases eta and kappa exchange with the polymerase delta holoenzyme to complete common fragile site synthesis
- PMID: 28605669
- PMCID: PMC9642814
- DOI: 10.1016/j.dnarep.2017.05.006
DNA polymerases eta and kappa exchange with the polymerase delta holoenzyme to complete common fragile site synthesis
Abstract
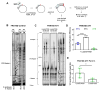
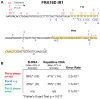
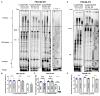
Common fragile sites (CFSs) are inherently unstable genomic loci that are recurrently altered in human tumor cells. Despite their instability, CFS are ubiquitous throughout the human genome and associated with large tumor suppressor genes or oncogenes. CFSs are enriched with repetitive DNA sequences, one feature postulated to explain why these loci are inherently difficult to replicate, and sensitive to replication stress. We have shown that specialized DNA polymerases (Pols) η and κ replicate CFS-derived sequences more efficiently than the replicative Pol δ. However, we lacked an understanding of how these enzymes cooperate to ensure efficient CFS replication. Here, we designed a model of lagging strand replication with RFC loaded PCNA that allows for maximal activity of the four-subunit human Pol δ holoenzyme, Pol η, and Pol κ in polymerase mixing assays. We discovered that Pol η and κ are both able to exchange with Pol δ stalled at repetitive CFS sequences, enhancing Normalized Replication Efficiency. We used this model to test the impact of PCNA mono-ubiquitination on polymerase exchange, and found no change in polymerase cooperativity in CFS replication compared with unmodified PCNA. Finally, we modeled replication stress in vitro using aphidicolin and found that Pol δ holoenzyme synthesis was significantly inhibited in a dose-dependent manner, preventing any replication past the CFS. Importantly, Pol η and κ were still proficient in rescuing this stalled Pol δ synthesis, which may explain, in part, the CFS instability phenotype of aphidicolin-treated Pol η and Pol κ-deficient cells. In total, our data support a model wherein Pol δ stalling at CFSs allows for free exchange with a specialized polymerase that is not driven by PCNA.
Keywords: AT microsatellite; DNA polymerase δ; DNA polymerase η; DNA polymerase κ; PCNA; TLS.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

