Diversity, classification and evolution of CRISPR-Cas systems
- PMID: 28605718
- PMCID: PMC5776717
- DOI: 10.1016/j.mib.2017.05.008
Diversity, classification and evolution of CRISPR-Cas systems
Abstract
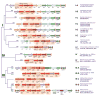
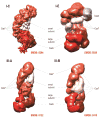
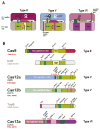
The bacterial and archaeal CRISPR-Cas systems of adaptive immunity show remarkable diversity of protein composition, effector complex structure, genome locus architecture and mechanisms of adaptation, pre-CRISPR (cr)RNA processing and interference. The CRISPR-Cas systems belong to two classes, with multi-subunit effector complexes in Class 1 and single-protein effector modules in Class 2. Concerted genomic and experimental efforts on comprehensive characterization of Class 2 CRISPR-Cas systems led to the identification of two new types and several subtypes. The newly characterized type VI systems are the first among the CRISPR-Cas variants to exclusively target RNA. Unexpectedly, in some of the class 2 systems, the effector protein is additionally responsible for the pre-crRNA processing. Comparative analysis of the effector complexes indicates that Class 2 systems evolved from mobile genetic elements on multiple, independent occasions.
Published by Elsevier Ltd.
Conflict of interest statement
Author declaration
We confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author.
Figures

References
-
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. - PubMed
-
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164(1–2):29–44. - PubMed
-
- Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2016 http://dx.doi.org/10.1016/j.cell.2016.10.044( - DOI - PubMed
-
- Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353(6299):aad5147. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

