Molecular epidemiology of Avian Rotaviruses Group A and D shed by different bird species in Nigeria
- PMID: 28606119
- PMCID: PMC5469043
- DOI: 10.1186/s12985-017-0778-5
Molecular epidemiology of Avian Rotaviruses Group A and D shed by different bird species in Nigeria
Abstract
Background: Avian rotaviruses (RVs) cause gastrointestinal diseases of birds worldwide. However, prevalence, diversity, epidemiology and phylogeny of RVs remain largely under-investigated in Africa.
Methods: Fecal samples from 349 birds (158 symptomatic, 107 asymptomatic and 84 birds without recorded health status) were screened by reverse transcription PCR to detect RV groups A and D (RVA and RVD). Partial gene sequences of VP4, VP6, VP7 and NSP4 for RVA, and of VP6 and VP7 for RVD were obtained and analyzed to infer phylogenetic relationship. Fisher's exact test and logistic regression were applied to identify factors potentially influencing virus shedding in chickens.
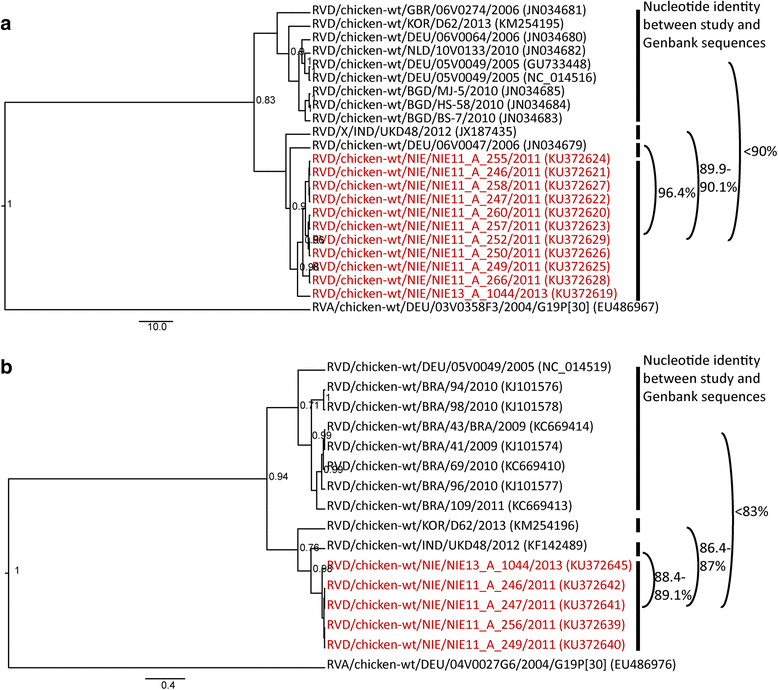
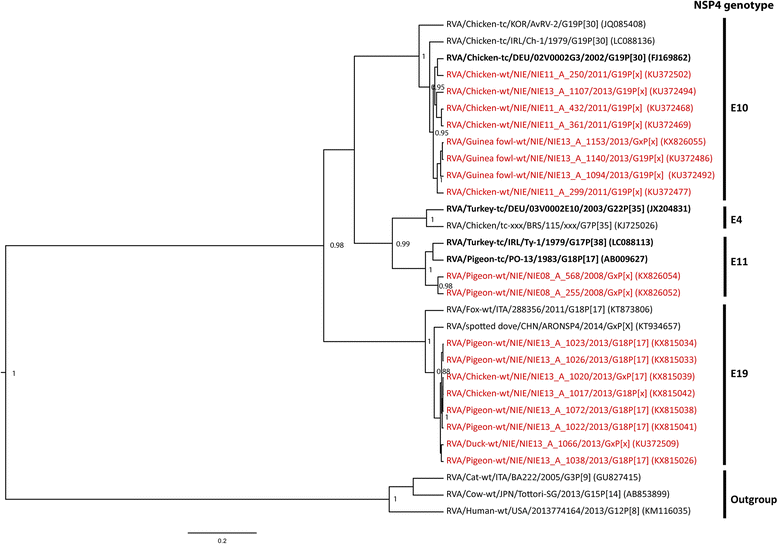
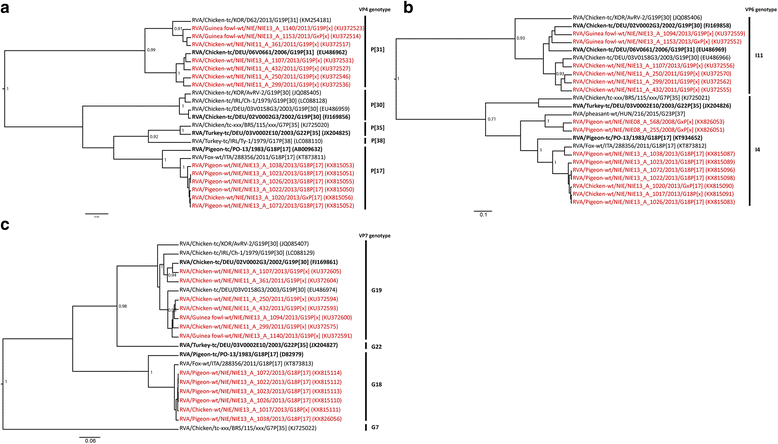
Results: A high prevalence of RVA (36.1%; 126/349) and RVD (31.8%; 111/349) shedding was revealed in birds. In chickens, RV shedding was age-dependent and highest RVD shedding rates were found in commercial farms. No negative health effect could be shown, and RVA and RVD shedding was significantly more likely in asymptomatic chickens: RVA/RVD were detected in 51.9/48.1% of the asymptomatic chickens, compared to 18.9/29.7% of the symptomatic chickens (p < 0.001/p = 0.01). First RVA sequences were obtained from mallard ducks (Anas platyrhynchos) and guinea fowls (Numida meleagris). Phylogenetic analyses illustrated the high genetic diversity of RVA and RVD in Nigerian birds and suggested cross-species transmission of RVA, especially at live bird markets. Indeed, RVA strains highly similar to a recently published fox rotavirus (RVA/Fox-tc/ITA/288356/2011/G18P[17]) and distantly related to other avian RVs were detected in different bird species, including pigeons, ducks, guinea fowls, quails and chickens.
Conclusion: This study provides new insights into epidemiology, diversity and classification of avian RVA and RVD in Nigeria. We show that cross-species transmission of host permissive RV strains occurs when different bird species are mixed.
Keywords: Avian rotaviruses; Epidemiology; Host permissiveness; Sub-Saharan Africa; Virus diversity.
Figures
References
-
- Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Volume 2. 5th edition. Philadelphia: Lippincott William & Wilkins; 2007.
-
- McNulty MS, Jones RC. Rotaviruses. In: Pattison, McMullin, Bradbury, Alexander, editors. Poultry diseases. 6th edition. Philadelphia: SAUNDERS Elsevier; 2008.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

