Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes
- PMID: 28606303
- PMCID: PMC5469618
- DOI: 10.7554/eLife.24634
Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes
Abstract
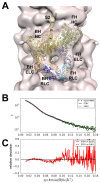
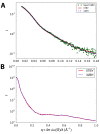
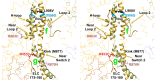
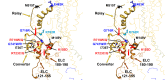
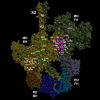
Cardiac β-myosin variants cause hypertrophic (HCM) or dilated (DCM) cardiomyopathy by disrupting sarcomere contraction and relaxation. The locations of variants on isolated myosin head structures predict contractility effects but not the prominent relaxation and energetic deficits that characterize HCM. During relaxation, pairs of myosins form interacting-heads motif (IHM) structures that with other sarcomere proteins establish an energy-saving, super-relaxed (SRX) state. Using a human β-cardiac myosin IHM quasi-atomic model, we defined interactions sites between adjacent myosin heads and associated protein partners, and then analyzed rare variants from 6112 HCM and 1315 DCM patients and 33,370 ExAC controls. HCM variants, 72% that changed electrostatic charges, disproportionately altered IHM interaction residues (expected 23%; HCM 54%, p=2.6×10-19; DCM 26%, p=0.66; controls 20%, p=0.23). HCM variant locations predict impaired IHM formation and stability, and attenuation of the SRX state - accounting for altered contractility, reduced diastolic relaxation, and increased energy consumption, that fully characterizes HCM pathogenesis.
Keywords: cardiomyopathy; diastolic heart disease; human; human biology; human mutation; medicine; myosin interacting-heads motif; super-relaxation; β-cardiac myosin.
Conflict of interest statement
JGS: is a founder and owns shares in Myokardia Inc., a startup company that is developing therapeutics that targets the sarcomere.
CES: is a founder and owns shares in Myokardia Inc., a startup company that is developing therapeutics that targets the sarcomere.
The other authors declare that no competing interests exist.
Figures










References
-
- Alamo L, Li XE, Espinoza-Fonseca LM, Pinto A, Thomas DD, Lehman W, Padrón R. Tarantula myosin free head regulatory light chain phosphorylation stiffens N-terminal extension, releasing it and blocking its docking back. Molecular bioSystems. 2015;11:2180–2189. doi: 10.1039/C5MB00163C. - DOI - PMC - PubMed
-
- Alamo L, Qi D, Wriggers W, Pinto A, Zhu J, Bilbao A, Gillilan RE, Hu S, Padrón R. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. Journal of Molecular Biology. 2016;428:1142–1164. doi: 10.1016/j.jmb.2016.01.027. - DOI - PMC - PubMed
-
- Alamo L, Wriggers W, Pinto A, Bártoli F, Salazar L, Zhao FQ, Craig R, Padrón R. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. Journal of Molecular Biology. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. - DOI - PMC - PubMed
-
- Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genetics in Medicine. 2015;17:880–888. doi: 10.1038/gim.2014.205. - DOI - PubMed

