TLR7/8 activation in neutrophils impairs immune complex phagocytosis through shedding of FcgRIIA
- PMID: 28606989
- PMCID: PMC5502427
- DOI: 10.1084/jem.20161512
TLR7/8 activation in neutrophils impairs immune complex phagocytosis through shedding of FcgRIIA
Abstract
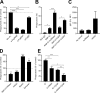
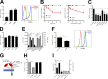
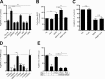
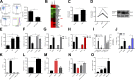
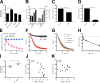
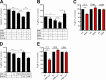
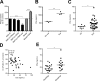
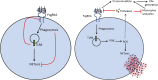
Neutrophils play a crucial role in host defense. However, neutrophil activation is also linked to autoimmune diseases such as systemic lupus erythematosus (SLE), where nucleic acid-containing immune complexes (IC) drive inflammation. The role of Toll-like receptor (TLR) signaling in processing of SLE ICs and downstream inflammatory neutrophil effector functions is not known. We observed that TLR7/8 activation leads to a furin-dependent proteolytic cleavage of the N-terminal part of FcgRIIA, shifting neutrophils away from phagocytosis of ICs toward the programmed form of necrosis, NETosis. TLR7/8-activated neutrophils promoted cleavage of FcgRIIA on plasmacytoid dendritic cells and monocytes, resulting in impaired overall clearance of ICs and increased complement C5a generation. Importantly, ex vivo derived activated neutrophils from SLE patients demonstrated a similar cleavage of FcgRIIA that was correlated with markers of disease activity, as well as complement activation. Therapeutic approaches aimed at blocking TLR7/8 activation would be predicted to increase phagocytosis of circulating ICs, while disarming their inflammatory potential.
© 2017 Lood et al.
Figures

References
-
- Anders A., Gilbert S., Garten W., Postina R., and Fahrenholz F.. 2001. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 15:1837–1839. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

