Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis
- PMID: 28607035
- PMCID: PMC5538628
- DOI: 10.15252/embr.201743947
Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis
Abstract
DNA-dependent activator of interferon regulatory factors/Z-DNA binding protein 1 (DAI/ZBP1) is a crucial sensor of necroptotic cell death induced by murine cytomegalovirus (MCMV) in its natural host. Here, we show that viral capsid transport to the nucleus and subsequent viral IE3-dependent early transcription are required for necroptosis. Necroptosis induction does not depend on input virion DNA or newly synthesized viral DNA A putative RNA-binding domain of DAI/ZBP1, Zα2, is required to sense virus and trigger necroptosis. Thus, MCMV IE3-dependent transcription from the viral genome plays a crucial role in activating DAI/ZBP1-dependent necroptosis. This implicates RNA transcripts generated by a large double-stranded DNA virus as a biologically relevant ligand for DAI/ZBP1 during natural viral infection.
Keywords: DAI/ZBP1; IE3; RIPK3; murine cytomegalovirus; necroptosis.
© 2017 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
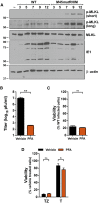
Immunoblot (IB) analysis to detect p‐MLKL, total MLKL, IE1, and β‐actin from SVEC4‐10 cells infected with bacmid‐derived K181 (WT) or M45mutRHIM MCMV at a multiplicity of infection (MOI) of 5.
Replication levels of WT MCMV in infected SVEC4‐10 cells 48 h post‐infection (h.p.i.) in the absence or presence of 200 μg/ml phosphonoformic acid (PFA; n = 3 biological replicates). Viral titers were determined by plaque assay.
Relative viability of SVEC4‐10 cells infected with M45mutRHIM compared to WT MCMV (MOI = 5) in the presence or absence of 200 μg/ml PFA (n = 3 biological replicates).
Relative viability of SVEC4‐10 cells treated with TNF (T) or TNF + ZVAD‐fmk (TZ) for 6 h in the presence or absence of 200 μg/ml PFA (n = 3 biological replicates).
IB analysis to detect IE1 and β‐actin from SVEC4‐10 cells infected with WT MCMV in the absence or presence of 1 μg/ml nocodazole (Noc).
Relative viability of SVEC4‐10 cells infected with M45mutRHIM compared to WT MCMV (MOI = 5) in the presence or absence 1 μg/ml Noc (n = 4 biological replicates).
IB analysis to detect p‐MLKL, total MLKL, IE1, and β‐actin from SVEC4‐10 cells infected with M45mutRHIM MCMV (MOI = 5) for 7 h in the presence or absence of 1 μg/ml Noc.
Relative viability of SVEC4‐10 cells treated with TNF (T), zVAD (Z), or TNF + zVAD‐fmk (TZ) for 6 h in the presence or absence of 1 μg/ml Noc (n = 3 biological replicates).
IB analysis to detect FLAG, RIPK3, and IE1 in subcellular fractions of 29‐11 cells stably reconstituted with FLAG‐epitope‐tagged WT DAI/ZBP1 and infected 7 h with WT or M45mutRHIM (MOI = 5). IB for β‐tubulin and histone H3 was used to assess quality of cytoplasmic and nuclear fractions, respectively.
Relative viability of SVEC4‐10 cells infected with M45mutRHIM compared to WT MCMV (MOI = 5) in the presence or absence of 100 μM Ciliobrevin‐D (CBD). **P < 0.01; two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).
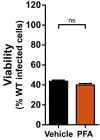
Relative viability of SVEC4‐10 cells treated with TNF (T), zVAD (Z), or TNF + zVAD‐fmk (TZ) for 6 h in the presence or absence of 100 μM CBD. n.s., not significant (P > 0.05) by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).
Representative results of sequencing and alignment of the DAI/ZBP1 exon 2 locus in 29‐11 cells. Amplicons generated from 29‐11 genomic DNA using ZBPSurveyor primers were cloned, and 11 independent clones sequenced.
Amino acid alignment of protein products predicted from the sequencing results in (C). Gray bars denote the boundaries of exon 1 (light) and exon 2 (dark) of DAI/ZBP1, and blue bar represents the defined Zα1 domain.
IB analysis to detect DAI/ZBP1, RIPK3, and β‐actin from SVEC‐10 parental and 29‐11 cell lines.

Schematic representation of timed α‐amanitin experiment.
Relative viability of SVEC4‐10 cells infected with M45mutRHIM compared to WT MCMV (MOI = 5) in the presence or absence of 50 μg/ml α‐amanitin according to the scheme in (A). ****P < 0.0001; *P < 0.05; n.s., not significant (P > 0.05) by one‐way ANOVA with Dunnett's multiple comparisons test, compared to untreated. Error bars indicate SD (n = 3 biological replicates).
IB analysis to detect p‐MLKL, total MLKL, and IE1 from SVEC4‐10 cells infected 7 h with WT or M45mutRHIM MCMV (MOI = 5).
Relative viability of SVEC4‐10 cells treated with TNF (T), zVAD (Z), or TNF + zVAD‐fmk (TZ) for 6 h in the presence or absence of 50 μg/ml α‐amanitin. n.s., not significant (P > 0.05) by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 independent biological replicates).
IB analysis to detect DAI/ZBP1, RIPK3, MLKL, IE1, and E1 from SVEC4‐10 cells infected with WT MCMV (MOI = 5) according to the scheme in (A).

Schematic representation of generation of recombinant WT‐IE3DD and M45mutRHIM IE3DD bacmids.
Schematic representation of destabilizing domain (DD) function. An IE3‐DD fusion is stabilized and functional in the presence of Shield‐1 (Shld). DD fusions are rapidly degraded in the absence of ligand.
Multistep replication levels of WT‐ and M45mutRHIM‐IE3DD viruses (MOI = 0.05) in NIH3T3 cells in the presence or absence of 1 μM Shld. Error bars indicate SD (n = 3 biological replicates).
Kinetics of cell death of SVEC4‐10 cells infected with WT‐ or M45mutRHIM‐IE3DD viruses (MOI = 5) in the presence or absence of 1 μM Shld, measured in real time by Sytox green incorporation. Error bars indicate SD (n = 4 biological replicates).
Relative viability of SVEC4‐10 cells infected with M45mutRHIM‐IE3DD compared to WT‐IE3DD (MOI = 5) in the presence or absence of 1 μM Shld. **P < 0.01; n.s., not significant (P > 0.05) by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).

RFLP analysis of parental WT, IE3.SK, IE3DD, parental M45mutRHIM, M45mutRHIM –IE3.SK, and M45mutRHIM‐IE3DD bacmids digested with EcoRI.
Agarose gel analysis of PCR amplicons generated from bacmids described in (A) with primers HS36 and HS37. Amplicons were digested with the indicated restriction endonuclease to diagnose the M45mutRHIM locus. un, undigested; B, BamHI; P, PvuII.
Agarose gel analysis of PCR amplicons generated from bacmids described in (A) with primers HS09 and HS10 to diagnose insertions at the end of IE3 exon 5.
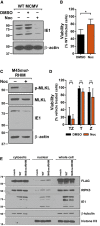
IB analysis to detect the DD‐domain (FKBP), E1, and β‐actin from NIH3T3 cells infected 13 h with parental or IE3DD WT or M45mutRHIM (RHIM) MCMV (MOI = 1) in the presence or absence of 1 μM Shield‐1.
Kinetics of cell death of 3T3‐SA cells infected with WT or M45mutRHIM (MOI = 5) in the presence or absence of 1 μM Shield‐1 (Shld), measured in real time by Sytox green incorporation. Error bars indicate SD (n = 4 biological replicates).
Multistep replication levels of parental WT and M45mutRHIM viruses (MOI = 0.05) in NIH3T3 cells in the presence or absence of 1 μM Shld. Error bars indicate SD (n = 3 biological replicates).
Relative viability of SVEC4‐10 cells infected with M45mutRHIM compared to WT MCMV in the absence or presence of 1 μM Shld. n.s., not significant (P > 0.05) by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).

- A
Schematic representation of the DAI/ZBP1 truncations and mutants. Blue ovals depict Z‐DNA‐binding domains, orange ovals represent RHIMS, and white ovals represent mutations. Numbers indicate amino acid positions.
- B
IB analysis to detect FLAG‐DAI/ZBP1, RIPK3, and β‐actin from 29‐11 cell line reconstituted by retroviral transduction with indicated DAI/ZBP1 mutant.
- C
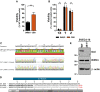
Relative viability of 29‐11 cell line reconstituted with indicated DAI/ZBP1 constructs infected with M45mutRHIM compared to WT MCMV (MOI = 5). ****P < 0.0001; n.s., not significant (P > 0.05) by one‐way ANOVA with Dunnett's multiple comparisons test, compared to WT. Error bars indicate standard deviation from the mean (SD; n = 3 biological replicates).
- D–F
Kinetics of cell death of indicated DAI/ZBP1‐reconstituted 29‐11 cell lines infected with M45mutRHIM MCMV (MOI = 5), measured in real time by Sytox green incorporation. Error bars indicate SD (n = 4 biological replicates).

Immunoprecipitation (IP) and IB analysis to detect FLAG‐DAI/ZBP1, myc‐RIPK3, and β‐actin from 293T cells transfected with vectors expressing 6myc‐RIPK3 and different 3XFLAG‐DAI/ZBP1 constructs, and subjected to IP with anti‐FLAG M2 agarose beads.
Cell death of DAI/ZBP1‐reconstituted 29‐11 cell lines infected with M45mutRHIM MCMV (MOI = 5); 6, 8, 10, and 12 h post‐infection times were extracted from Fig 5D–F and replotted for clarity. ****P < 0.0001; ***P < 0.001; *P < 0.05; n.s., not significant (P > 0.05) by one‐way ANOVA with Dunnett's multiple comparisons test, compared to EV. Error bars indicate standard deviation from the mean (SD; n = 4 biological replicates).
Relative viability of HT‐29 cells with or without hDAI expression infected with M45mutRHIM compared to WT MCMV. ***P < 0.001 by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).
IB analysis to detect p‐MLKL, total MLKL, and β‐actin from HT‐29 cells with or without hDAI expression infected with WT or M45mutRHIM for 10 h.
Relative viability of 29‐11 cells reconstituted with empty vector (EV) or DAI‐ΔZα1mutZα2 infected with M45mutRHIM compared to WT MCMV. ***P < 0.001 by two‐tailed unpaired Student's t‐test. Error bars indicate SD (n = 3 biological replicates).
Comment in
-
Live and let die: ZBP1 senses viral and cellular RNAs to trigger necroptosis.EMBO J. 2017 Sep 1;36(17):2470-2472. doi: 10.15252/embj.201797845. Epub 2017 Aug 18. EMBO J. 2017. PMID: 28821535 Free PMC article.
Similar articles
-
DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA.Cell Host Microbe. 2012 Mar 15;11(3):290-7. doi: 10.1016/j.chom.2012.01.016. Cell Host Microbe. 2012. PMID: 22423968 Free PMC article.
-
Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1.Cell Death Dis. 2018 Jul 26;9(8):816. doi: 10.1038/s41419-018-0868-3. Cell Death Dis. 2018. PMID: 30050136 Free PMC article.
-
RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation.Nature. 2016 Dec 1;540(7631):124-128. doi: 10.1038/nature20558. Epub 2016 Nov 7. Nature. 2016. PMID: 27819681 Free PMC article.
-
Viral Z-RNA triggers ZBP1-dependent cell death.Curr Opin Virol. 2021 Dec;51:134-140. doi: 10.1016/j.coviro.2021.10.004. Epub 2021 Oct 21. Curr Opin Virol. 2021. PMID: 34688984 Free PMC article. Review.
-
Z-DNA binding protein 1 orchestrates innate immunity and inflammatory cell death.Cytokine Growth Factor Rev. 2024 Jun;77:15-29. doi: 10.1016/j.cytogfr.2024.03.005. Epub 2024 Mar 26. Cytokine Growth Factor Rev. 2024. PMID: 38548490 Review.
Cited by
-
ZBP1 not RIPK1 mediates tumor necroptosis in breast cancer.Nat Commun. 2021 May 11;12(1):2666. doi: 10.1038/s41467-021-23004-3. Nat Commun. 2021. PMID: 33976222 Free PMC article.
-
Mechanisms of TNF-independent RIPK3-mediated cell death.Biochem J. 2022 Oct 14;479(19):2049-2062. doi: 10.1042/BCJ20210724. Biochem J. 2022. PMID: 36240069 Free PMC article. Review.
-
Emerging Role of ZBP1 in Z-RNA Sensing, Influenza Virus-Induced Cell Death, and Pulmonary Inflammation.mBio. 2022 Jun 28;13(3):e0040122. doi: 10.1128/mbio.00401-22. Epub 2022 May 19. mBio. 2022. PMID: 35587190 Free PMC article. Review.
-
Cell Killing by Reovirus: Mechanisms and Consequences.Curr Top Microbiol Immunol. 2023;442:133-153. doi: 10.1007/82_2020_225. Curr Top Microbiol Immunol. 2023. PMID: 32986138 Free PMC article. Review.
-
RIPK1 is essential for Herpes Simplex Virus-triggered ZBP1-dependent necroptosis in human cells.bioRxiv [Preprint]. 2024 Sep 19:2024.09.17.613393. doi: 10.1101/2024.09.17.613393. bioRxiv. 2024. PMID: 39345610 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

