Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading
- PMID: 28607053
- PMCID: PMC5495254
- DOI: 10.1073/pnas.1702383114
Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading
Abstract
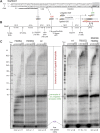
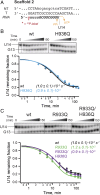
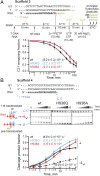
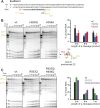
The active site of multisubunit RNA polymerases (RNAPs) is highly conserved from humans to bacteria. This single site catalyzes both nucleotide addition required for RNA transcript synthesis and excision of incorrect nucleotides after misincorporation as a proofreading mechanism. Phosphoryl transfer and proofreading hydrolysis are controlled in part by a dynamic RNAP component called the trigger loop (TL), which cycles between an unfolded loop and an α-helical hairpin [trigger helices (TH)] required for rapid nucleotide addition. The precise roles of the TL/TH in RNA synthesis and hydrolysis remain unclear. An invariant histidine residue has been proposed to function in the TH form as a general acid in RNA synthesis and as a general base in RNA hydrolysis. The effects of conservative, nonionizable substitutions of the TL histidine (or a neighboring TL arginine conserved in bacteria) have not yet been rigorously tested. Here, we report that glutamine substitutions of these residues, which preserve polar interactions but are incapable of acid-base chemistry, had little effect on either phosphoryl transfer or proofreading hydrolysis by Escherichia coli RNAP. The TL substitutions did, however, affect the backtracking of RNAP necessary for proofreading and potentially the reactivity of the backtracked nucleotide. We describe a unifying model for the function of the RNAP TL, which reconciles available data and our results for representative RNAPs. This model explains diverse effects of the TL basic residues on catalysis through their effects on positioning reactants for phosphoryl transfer and easing barriers to transcript backtracking, rather than as acid-base catalysts.
Keywords: RNA hydrolysis; acid–base catalysis; proofreading; transcription; trigger loop.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. - PubMed
-
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. - PubMed
-
- Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

