A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
- PMID: 28607067
- PMCID: PMC5495272
- DOI: 10.1073/pnas.1705689114
A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Abstract
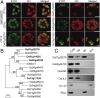
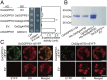
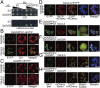
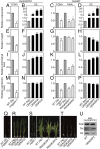
In plants, geranylgeranyl diphosphate (GGPP) is produced by plastidic GGPP synthase (GGPPS) and serves as a precursor for vital metabolic branches, including chlorophyll, carotenoid, and gibberellin biosynthesis. However, molecular mechanisms regulating GGPP allocation among these biosynthetic pathways localized in the same subcellular compartment are largely unknown. We found that rice contains only one functionally active GGPPS, OsGGPPS1, in chloroplasts. A functionally active homodimeric enzyme composed of two OsGGPPS1 subunits is located in the stroma. In thylakoid membranes, however, the GGPPS activity resides in a heterodimeric enzyme composed of one OsGGPPS1 subunit and GGPPS recruiting protein (OsGRP). OsGRP is structurally most similar to members of the geranyl diphosphate synthase small subunit type II subfamily. In contrast to members of this subfamily, OsGRP enhances OsGGPPS1 catalytic efficiency and specificity of GGPP production on interaction with OsGGPPS1. Structural biology and protein interaction analyses demonstrate that affinity between OsGRP and OsGGPPS1 is stronger than between two OsGGPPS1 molecules in homodimers. OsGRP determines OsGGPPS1 suborganellar localization and directs it to a large protein complex in thylakoid membranes, consisting of geranylgeranyl reductase (OsGGR), light-harvesting-like protein 3 (OsLIL3), protochlorophyllide oxidoreductase (OsPORB), and chlorophyll synthase (OsCHLG). Taken together, genetic and biochemical analyses suggest OsGRP functions in recruiting OsGGPPS1 from the stroma toward thylakoid membranes, thus providing a mechanism to control GGPP flux toward chlorophyll biosynthesis.
Keywords: chlorophyll biosynthesis; geranylgeranyl diphosphate synthase; protein complex formation; rice; terpenoids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hemmerlin A, Harwood JL, Bach TJ. A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res. 2012;51:95–148. - PubMed
-
- Tholl D. Biosynthesis and biological functions of terpenoids in plants. In: Schrader J, Bohlmann J, editors. Advances in Biochemical Engineering/Biotechnology. Vol 148. Springer; New York: 2015. pp. 63–106. - PubMed
-
- Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. - PubMed
-
- Ashour M, Wink M, Gershenzon J. Biochemistry of terpenoids: Monoterpenes, sesquiterpenes and diterpenes. In: Wink M, editor. Annual Plant Reviews. 2nd Ed. Vol 40. Blackwell; Singapore: 2010. pp. 258–303.
-
- Wang KC, Ohnuma S. Isoprenyl diphosphate synthases. Biochim Biophys Acta. 2000;1529:33–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

