NMR structure of the Bacillus cereus hemolysin II C-terminal domain reveals a novel fold
- PMID: 28607368
- PMCID: PMC5468326
- DOI: 10.1038/s41598-017-02917-4
NMR structure of the Bacillus cereus hemolysin II C-terminal domain reveals a novel fold
Abstract
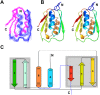
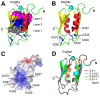
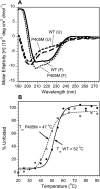
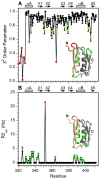
In addition to multiple virulence factors, Bacillus cereus a pathogen that causes food poisoning and life-threatening wound infections, secretes the pore-forming toxin hemolysin II (HlyII). The HlyII toxin has a unique 94 amino acid C-terminal domain (HlyIIC). HlyIIC exhibits splitting of NMR resonances due to cis/trans isomerization of a single proline near the C-terminus. To overcome heterogeneity, we solved the structure of P405M-HlyIIC, a mutant that exclusively stabilizes the trans state. The NMR structure of HlyIIC reveals a novel fold, consisting of two subdomains αA-β1-β2 and β3-β4-αB-β5, that come together in a barrel-like structure. The barrel core is fastened by three layers of hydrophobic residues. The barrel end opposite the HlyIIC-core has a positively charged surface, that by binding negatively charged moieties on cellular membranes, may play a role in target-cell surface recognition or stabilization of the heptameric pore complex. In the WT domain, dynamic flexibility occurs at the N-terminus and the first α-helix that connects the HlyIIC domain to the HlyII-core structure. In the destabilizing P405M mutant, increased flexibility is evident throughout the first subdomain, suggesting that the HlyIIC structure may have arisen through gene fusion.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
The Food Poisoning Toxins of Bacillus cereus.Toxins (Basel). 2021 Jan 28;13(2):98. doi: 10.3390/toxins13020098. Toxins (Basel). 2021. PMID: 33525722 Free PMC article. Review.
-
Protein yoga: Conformational versatility of the Hemolysin II C-terminal domain detailed by NMR structures for multiple states.Protein Sci. 2021 May;30(5):990-1005. doi: 10.1002/pro.4066. Epub 2021 Mar 30. Protein Sci. 2021. PMID: 33733504 Free PMC article.
-
NMR assignments for the cis and trans forms of the hemolysin II C-terminal domain.Biomol NMR Assign. 2014 Oct;8(2):419-23. doi: 10.1007/s12104-013-9530-2. Biomol NMR Assign. 2014. PMID: 24234348
-
A Monoclonal Antibody against the C-Terminal Domain of Bacillus cereus Hemolysin II Inhibits HlyII Cytolytic Activity.Toxins (Basel). 2020 Dec 19;12(12):806. doi: 10.3390/toxins12120806. Toxins (Basel). 2020. PMID: 33352744 Free PMC article.
-
The pore-forming haemolysins of bacillus cereus: a review.Toxins (Basel). 2013 Jun 7;5(6):1119-39. doi: 10.3390/toxins5061119. Toxins (Basel). 2013. PMID: 23748204 Free PMC article. Review.
Cited by
-
The Food Poisoning Toxins of Bacillus cereus.Toxins (Basel). 2021 Jan 28;13(2):98. doi: 10.3390/toxins13020098. Toxins (Basel). 2021. PMID: 33525722 Free PMC article. Review.
-
Utilizing Extraepitopic Amino Acid Substitutions to Define Changes in the Accessibility of Conformational Epitopes of the Bacillus cereus HlyII C-Terminal Domain.Int J Mol Sci. 2023 Nov 17;24(22):16437. doi: 10.3390/ijms242216437. Int J Mol Sci. 2023. PMID: 38003626 Free PMC article.
-
Protein yoga: Conformational versatility of the Hemolysin II C-terminal domain detailed by NMR structures for multiple states.Protein Sci. 2021 May;30(5):990-1005. doi: 10.1002/pro.4066. Epub 2021 Mar 30. Protein Sci. 2021. PMID: 33733504 Free PMC article.
-
The Temperature Dependence of Hydrogen Bonds Is More Uniform in Stable Proteins: An Analysis of NMR h3JNC' Couplings in Four Different Protein Structures.Molecules. 2024 Jun 21;29(13):2950. doi: 10.3390/molecules29132950. Molecules. 2024. PMID: 38998901 Free PMC article.
-
Monoclonal Antibody HlyIIC‑15 to C-End Domain HlyII B. cereus Interacts with the Trombin Recognition Site.Russ J Bioorg Chem. 2020;46(6):1214-1220. doi: 10.1134/S1068162020060382. Epub 2020 Dec 28. Russ J Bioorg Chem. 2020. PMID: 33390685 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

