Principles of early human development and germ cell program from conserved model systems
- PMID: 28607482
- PMCID: PMC5473469
- DOI: 10.1038/nature22812
Principles of early human development and germ cell program from conserved model systems
Abstract
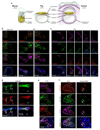
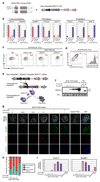
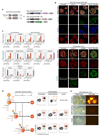
Human primordial germ cells (hPGCs), the precursors of sperm and eggs, originate during weeks 2-3 of early post-implantation development. Using in vitro models of hPGC induction, recent studies have suggested that there are marked mechanistic differences in the specification of human and mouse PGCs. This may be due in part to the divergence in their pluripotency networks and early post-implantation development. As early human embryos are not accessible for direct study, we considered alternatives including porcine embryos that, as in humans, develop as bilaminar embryonic discs. Here we show that porcine PGCs originate from the posterior pre-primitive-streak competent epiblast by sequential upregulation of SOX17 and BLIMP1 in response to WNT and BMP signalling. We use this model together with human and monkey in vitro models simulating peri-gastrulation development to show the conserved principles of epiblast development for competency for primordial germ cell fate. This process is followed by initiation of the epigenetic program and regulated by a balanced SOX17-BLIMP1 gene dosage. Our combinatorial approach using human, porcine and monkey in vivo and in vitro models provides synthetic insights into early human development.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

