Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer
- PMID: 28607485
- PMCID: PMC5538883
- DOI: 10.1038/nature22341
Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer
Abstract
The mutant form of the GTPase KRAS is a key driver of pancreatic cancer but remains a challenging therapeutic target. Exosomes are extracellular vesicles generated by all cells, and are naturally present in the blood. Here we show that enhanced retention of exosomes, compared to liposomes, in the circulation of mice is likely due to CD47-mediated protection of exosomes from phagocytosis by monocytes and macrophages. Exosomes derived from normal fibroblast-like mesenchymal cells were engineered to carry short interfering RNA or short hairpin RNA specific to oncogenic KrasG12D, a common mutation in pancreatic cancer. Compared to liposomes, the engineered exosomes (known as iExosomes) target oncogenic KRAS with an enhanced efficacy that is dependent on CD47, and is facilitated by macropinocytosis. Treatment with iExosomes suppressed cancer in multiple mouse models of pancreatic cancer and significantly increased overall survival. Our results demonstrate an approach for direct and specific targeting of oncogenic KRAS in tumours using iExosomes.
Conflict of interest statement
MD Anderson Cancer Center and RK hold patents in the area of exosome biology and are licensed to Codiak Biosciences Inc. MD Anderson Cancer Center and RK are stock equity holders in Codiak Biosciences Inc. RK receives research support from Codiak Biosciences Inc. and serves as a member of the board of directors. VSL served once as a paid consultant for Codiak Biosciences Inc.
Figures










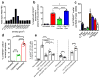

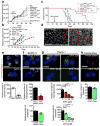

Comment in
-
Pancreatic cancer: iExosomes target the 'undruggable'.Nat Rev Cancer. 2017 Aug;17(8):452-453. doi: 10.1038/nrc.2017.54. Epub 2017 Jun 30. Nat Rev Cancer. 2017. PMID: 28663589 No abstract available.
-
Cancer: RNA-based approaches target KRAS.Nat Rev Drug Discov. 2017 Jul 31;16(8):529. doi: 10.1038/nrd.2017.137. Nat Rev Drug Discov. 2017. PMID: 28757631 No abstract available.
-
Pancreatic cancer: Exosomes for targeting KRAS in the treatment of pancreatic cancer.Nat Rev Gastroenterol Hepatol. 2017 Nov;14(11):636-638. doi: 10.1038/nrgastro.2017.113. Epub 2017 Sep 6. Nat Rev Gastroenterol Hepatol. 2017. PMID: 28874835 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

