Single-molecule analysis of ligand efficacy in β2AR-G-protein activation
- PMID: 28607487
- PMCID: PMC5502743
- DOI: 10.1038/nature22354
Single-molecule analysis of ligand efficacy in β2AR-G-protein activation
Abstract
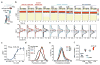
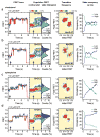
G-protein-coupled receptor (GPCR)-mediated signal transduction is central to human physiology and disease intervention, yet the molecular mechanisms responsible for ligand-dependent signalling responses remain poorly understood. In class A GPCRs, receptor activation and G-protein coupling entail outward movements of transmembrane helix 6 (TM6). Here, using single-molecule fluorescence resonance energy transfer imaging, we examine TM6 movements in the β2 adrenergic receptor (β2AR) upon exposure to orthosteric ligands with different efficacies, in the absence and presence of the Gs heterotrimer. We show that partial and full agonists differentially affect TM6 motions to regulate the rate at which GDP-bound β2AR-Gs complexes are formed and the efficiency of nucleotide exchange leading to Gs activation. These data also reveal transient nucleotide-bound β2AR-Gs species that are distinct from known structures, and provide single-molecule perspectives on the allosteric link between ligand- and nucleotide-binding pockets that shed new light on the G-protein activation mechanism.
Conflict of interest statement
The authors declare competing financial interest: details accompany the full-text HTML version of the paper (nature.com).
Figures








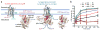



References
-
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

