Stability and function of regulatory T cells expressing the transcription factor T-bet
- PMID: 28607488
- PMCID: PMC5482236
- DOI: 10.1038/nature22360
Stability and function of regulatory T cells expressing the transcription factor T-bet
Erratum in
-
Corrigendum: Stability and function of regulatory T cells expressing the transcription factor T-bet.Nature. 2017 Sep 20;550(7674):142. doi: 10.1038/nature24013. Nature. 2017. PMID: 28953866
Abstract
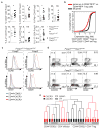
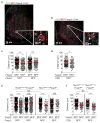
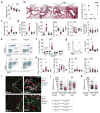
Adaptive immune responses are tailored to different types of pathogens through differentiation of naive CD4 T cells into functionally distinct subsets of effector T cells (T helper 1 (TH1), TH2, and TH17) defined by expression of the key transcription factors T-bet, GATA3, and RORγt, respectively. Regulatory T (Treg) cells comprise a distinct anti-inflammatory lineage specified by the X-linked transcription factor Foxp3 (refs 2, 3). Paradoxically, some activated Treg cells express the aforementioned effector CD4 T cell transcription factors, which have been suggested to provide Treg cells with enhanced suppressive capacity. Whether expression of these factors in Treg cells-as in effector T cells-is indicative of heterogeneity of functionally discrete and stable differentiation states, or conversely may be readily reversible, is unknown. Here we demonstrate that expression of the TH1-associated transcription factor T-bet in mouse Treg cells, induced at steady state and following infection, gradually becomes highly stable even under non-permissive conditions. Loss of function or elimination of T-bet-expressing Treg cells-but not of T-bet expression in Treg cells-resulted in severe TH1 autoimmunity. Conversely, following depletion of T-bet- Treg cells, the remaining T-bet+ cells specifically inhibited TH1 and CD8 T cell activation consistent with their co-localization with T-bet+ effector T cells. These results suggest that T-bet+ Treg cells have an essential immunosuppressive function and indicate that Treg cell functional heterogeneity is a critical feature of immunological tolerance.
Conflict of interest statement
The authors declare no completing financial interests.
Figures












Comment in
-
Regulatory T cells: Subset-specific suppression.Nat Rev Immunol. 2017 Jul;17(7):401. doi: 10.1038/nri.2017.68. Epub 2017 Jun 19. Nat Rev Immunol. 2017. PMID: 28627521 No abstract available.
References
-
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–336. - PubMed
-
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. - PubMed
-
- Ohnmacht C, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

