Computational modeling of the bat HKU4 coronavirus 3CLpro inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus
- PMID: 28608547
- PMCID: PMC7166879
- DOI: 10.1002/jmr.2644
Computational modeling of the bat HKU4 coronavirus 3CLpro inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus
Abstract
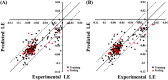
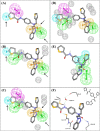
The Middle East respiratory syndrome coronavirus (MERS-CoV) is an emerging virus that poses a major challenge to clinical management. The 3C-like protease (3CLpro ) is essential for viral replication and thus represents a potential target for antiviral drug development. Presently, very few data are available on MERS-CoV 3CLpro inhibition by small molecules. We conducted extensive exploration of the pharmacophoric space of a recently identified set of peptidomimetic inhibitors of the bat HKU4-CoV 3CLpro . HKU4-CoV 3CLpro shares high sequence identity (81%) with the MERS-CoV enzyme and thus represents a potential surrogate model for anti-MERS drug discovery. We used 2 well-established methods: Quantitative structure-activity relationship (QSAR)-guided modeling and docking-based comparative intermolecular contacts analysis. The established pharmacophore models highlight structural features needed for ligand recognition and revealed important binding-pocket regions involved in 3CLpro -ligand interactions. The best models were used as 3D queries to screen the National Cancer Institute database for novel nonpeptidomimetic 3CLpro inhibitors. The identified hits were tested for HKU4-CoV and MERS-CoV 3CLpro inhibition. Two hits, which share the phenylsulfonamide fragment, showed moderate inhibitory activity against the MERS-CoV 3CLpro and represent a potential starting point for the development of novel anti-MERS agents. To the best of our knowledge, this is the first pharmacophore modeling study supported by in vitro validation on the MERS-CoV 3CLpro .
Highlights: MERS-CoV is an emerging virus that is closely related to the bat HKU4-CoV. 3CLpro is a potential drug target for coronavirus infection. HKU4-CoV 3CLpro is a useful surrogate model for the identification of MERS-CoV 3CLpro enzyme inhibitors. dbCICA is a very robust modeling method for hit identification. The phenylsulfonamide scaffold represents a potential starting point for MERS coronavirus 3CLpro inhibitors development.
Keywords: 3CLpro inhibitors; MERS; coronavirus; dbCICA; pharmacophore modeling.
Copyright © 2017 John Wiley & Sons, Ltd.
Figures




References
-
- Dudley JP. Middle East respiratory syndrome coronavirus. Global Virus Network [Internet]. 2014.
-
- WHO . Middle East respiratory syndrome coronavirus (MERS‐CoV) WHO MERS‐CoV global summary and risk assessment 2016.
-
- Collier L, Oxford J, Kellam P. Human Virology. Fifth ed. Oxford: Oxford University Press; 2016.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

