A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis
- PMID: 28609658
- PMCID: PMC5649440
- DOI: 10.1016/j.ccell.2017.05.007
A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis
Abstract
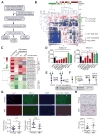
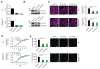
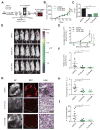
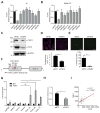
Association of aberrant glycosylation with melanoma progression is based mainly on analyses of cell lines. Here we present a systems-based study of glycomic changes and corresponding enzymes associated with melanoma metastasis in patient samples. Upregulation of core fucosylation (FUT8) and downregulation of α-1,2 fucosylation (FUT1, FUT2) were identified as features of metastatic melanoma. Using both in vitro and in vivo studies, we demonstrate FUT8 is a driver of melanoma metastasis which, when silenced, suppresses invasion and tumor dissemination. Glycoprotein targets of FUT8 were enriched in cell migration proteins including the adhesion molecule L1CAM. Core fucosylation impacted L1CAM cleavage and the ability of L1CAM to support melanoma invasion. FUT8 and its targets represent therapeutic targets in melanoma metastasis.
Keywords: FUT8; L1CAM; core fucosylation; glycomics; glycosylation; lectin array; lectin microarray; metastasis; metastatic melanoma; primary melanoma.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Aberrant Glycosylation in Cancer: A Novel Molecular Mechanism Controlling Metastasis.Cancer Cell. 2017 Jun 12;31(6):733-735. doi: 10.1016/j.ccell.2017.05.012. Cancer Cell. 2017. PMID: 28609653
References
-
- Altevogt P, Doberstein K, Fogel M. L1CAM in human cancer. International journal of cancer. 2016;138:1565–1576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

