The lost language of the RNA World
- PMID: 28611182
- PMCID: PMC5789781
- DOI: 10.1126/scisignal.aam8812
The lost language of the RNA World
Abstract
The possibility of an RNA World is based on the notion that life on Earth passed through a primitive phase without proteins, a time when all genomes and enzymes were composed of ribonucleic acids. Numerous apparent vestiges of this ancient RNA World remain today, including many nucleotide-derived coenzymes, self-processing ribozymes, metabolite-binding riboswitches, and even ribosomes. Many of the most common signaling molecules and second messengers used by modern organisms are also formed from RNA nucleotides or their precursors. For example, nucleotide derivatives such as cAMP, ppGpp, and ZTP, as well as the cyclic dinucleotides c-di-GMP and c-di-AMP, are intimately involved in signaling diverse physiological or metabolic changes in bacteria and other organisms. We describe the potential diversity of this "lost language" of the RNA World and speculate on whether additional components of this ancient communication machinery might remain hidden though still very much relevant to modern cells.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

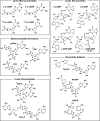

References
-
- Woese C. The genetic code: the molecular basis for genetic expression. Harper & Row. 1967
-
- Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–379. - PubMed
-
- Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38:381–393. - PubMed
-
- Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618.
-
- Joyce GF. The rise and fall of the RNA world. New Biol. 1991;3:399–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

