Microbial Community and Functional Structure Significantly Varied among Distinct Types of Paddy Soils But Responded Differently along Gradients of Soil Depth Layers
- PMID: 28611747
- PMCID: PMC5447084
- DOI: 10.3389/fmicb.2017.00945
Microbial Community and Functional Structure Significantly Varied among Distinct Types of Paddy Soils But Responded Differently along Gradients of Soil Depth Layers
Abstract
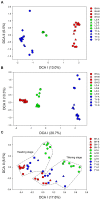
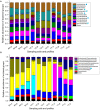
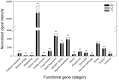
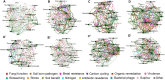
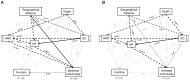
Paddy rice fields occupy broad agricultural area in China and cover diverse soil types. Microbial community in paddy soils is of great interest since many microorganisms are involved in soil functional processes. In the present study, Illumina Mi-Seq sequencing and functional gene array (GeoChip 4.2) techniques were combined to investigate soil microbial communities and functional gene patterns across the three soil types including an Inceptisol (Binhai), an Oxisol (Leizhou), and an Ultisol (Taoyuan) along four profile depths (up to 70 cm in depth) in mesocosm incubation columns. Detrended correspondence analysis revealed that distinctly differentiation in microbial community existed among soil types and profile depths, while the manifest variance in functional structure was only observed among soil types and two rice growth stages, but not across profile depths. Along the profile depth within each soil type, Acidobacteria, Chloroflexi, and Firmicutes increased whereas Cyanobacteria, β-proteobacteria, and Verrucomicrobia declined, suggesting their specific ecophysiological properties. Compared to bacterial community, the archaeal community showed a more contrasting pattern with the predominant groups within phyla Euryarchaeota, Thaumarchaeota, and Crenarchaeota largely varying among soil types and depths. Phylogenetic molecular ecological network (pMEN) analysis further indicated that the pattern of bacterial and archaeal communities interactions changed with soil depth and the highest modularity of microbial community occurred in top soils, implying a relatively higher system resistance to environmental change compared to communities in deeper soil layers. Meanwhile, microbial communities had higher connectivity in deeper soils in comparison with upper soils, suggesting less microbial interaction in surface soils. Structure equation models were developed and the models indicated that pH was the most representative characteristics of soil type and identified as the key driver in shaping both bacterial and archaeal community structure, but did not directly affect microbial functional structure. The distinctive pattern of microbial taxonomic and functional composition along soil profiles implied functional redundancy within these paddy soils.
Keywords: GeoChip; Mi-Seq sequencing; microbial community; network analysis; paddy soil; soil profile; soil type.
Figures

References
-
- Anderson D. (1988). The effect of parent material and soil development on nutrient cycling in temperate ecosystems. Biogeochemistry 5 71–97. 10.1007/BF02180318 - DOI
-
- Asghar H. N., Setia R., Marschner P. (2012). Community composition and activity of microbes from saline soils and non-saline soils respond similarly to changes in salinity. Soil Biol. Biochem. 47 175–178. 10.1016/j.soilbio.2012.01.002 - DOI
-
- Bai R., Xi D., He J.-Z., Hu H.-W., Fang Y.-T., Zhang L.-M. (2015). Activity, abundance and community structure of anammox bacteria along depth profiles in three different paddy soils. Soil Biol. Biochem. 91 212–221. 10.1016/j.soilbio.2015.08.040 - DOI
-
- Bastian M., Heymann S. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, CA.
LinkOut - more resources
Full Text Sources
Other Literature Sources

