A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus
- PMID: 28613431
- PMCID: PMC5570653
- DOI: 10.1111/mmi.13731
A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus
Abstract
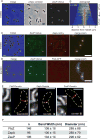
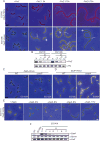
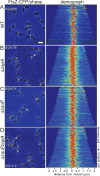
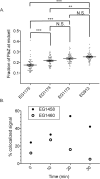
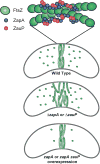
The cytoskeletal GTPase FtsZ assembles at midcell, recruits the division machinery and directs envelope invagination for bacterial cytokinesis. ZapA, a conserved FtsZ-binding protein, promotes Z-ring stability and efficient division through a mechanism that is not fully understood. Here, we investigated the function of ZapA in Caulobacter crescentus. We found that ZapA is encoded in an operon with a small coiled-coil protein we named ZauP. ZapA and ZauP co-localized at the division site and were each required for efficient division. ZapA interacted directly with both FtsZ and ZauP. Neither ZapA nor ZauP influenced FtsZ dynamics or bundling, in vitro, however. Z-rings were diffuse in cells lacking zapA or zauP and, conversely, FtsZ was enriched at midcell in cells overproducing ZapA and ZauP. Additionally, FtsZ persisted at the poles longer when ZapA and ZauP were overproduced, and frequently colocalized with MipZ, a negative regulator of FtsZ polymerization. We propose that ZapA and ZauP promote efficient cytokinesis by stabilizing the midcell Z-ring through a bundling-independent mechanism. The zauPzapA operon is present in diverse Gram-negative bacteria, indicating a common mechanism for Z-ring assembly.
© 2017 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that no conflicts of interest exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

