Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam
- PMID: 28614360
- PMCID: PMC5484541
- DOI: 10.1371/journal.pntd.0005628
Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam
Abstract
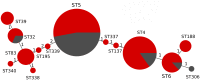
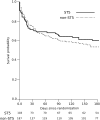
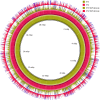
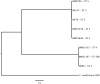
The vast burden of cryptococcal meningitis occurs in immunosuppressed patients, driven by HIV, and is caused by Cryptococcus neoformans var. grubii. We previously reported cryptococcal meningitis in Vietnam arising atypically in HIV uninfected, apparently immunocompetent patients, caused by a single amplified fragment length polymorphism (AFLP) cluster of C. neoformans var. grubii (VNIγ). This variant was less common in HIV infected individuals; it remains unclear why this lineage is associated with apparently immunocompetent patients. To study this host tropism we aimed to further our understanding of clinical phenotype and genomic variation within Vietnamese C. neoformans var. grubii. After performing MLST on C. neoformans clinical isolates we identified 14 sequence types (STs); ST5 correlated with the VNIγ cluster. We next compared clinical phenotype by lineage and found HIV infected patients with cryptococcal meningitis caused by ST5 organisms were significantly more likely to have lymphadenopathy (11% vs. 4%, p = 0.05 Fisher's exact test) and higher blood lymphocyte count (median 0.76 versus 0.55 X109 cells/L, p = 0.001, Kruskal-Wallis test). Furthermore, survivors of ST5 infections had evidence of worse disability outcomes at 70 days (72.7% (40/55) in ST5 infections versus 57.1% (52/91) non-ST5 infections (OR 2.11, 95%CI 1.01 to 4.41), p = 0.046). To further investigate the relationship between strain and disease phenotype we performed genome sequencing on eight Vietnamese C. neoformans var. grubii. Eight genome assemblies exhibited >99% nucleotide sequence identity and we identified 165 kbp of lineage specific to Vietnamese isolates. ST5 genomes harbored several strain specific regions, incorporating 19 annotated coding sequences and eight hypothetical proteins. These regions included a phenolic acid decarboxylase, a DEAD-box ATP-dependent RNA helicase 26, oxoprolinases, a taurine catabolism dioxygenase, a zinc finger protein, membrane transport proteins and various drug transporters. Our work outlines the complexity of genomic pathogenicity in cryptococcal infections and identifies a number of gene candidates that may aid the disaggregation of the pathways associated with the pathogenesis of Cryptococcus neoformans var. grubii.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids. 2009;23(4):525–30. doi: 10.1097/QAD.0b013e328322ffac - DOI - PubMed
-
- Casadevall A, Perfect JR. Cryptococcus neoformans. 1 ed Washington: American Society for Microbiology Press; 1998. 1998. 541 p.
-
- Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. ClinInfectDis. 2000;31(2):499–508. - PubMed
-
- Dromer F, Mathoulin S, Dupont B, Laporte A. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993). French Cryptococcosis Study Group. ClinInfectDis. 1996;23(1):82–90. - PubMed
-
- Chau TT, Mai NH, Phu NH, Nghia HD, Chuong LV, Sinh DX, et al. A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam—high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect Dis. 2010;10:199 doi: 10.1186/1471-2334-10-199 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

