Genome-wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements
- PMID: 28614382
- PMCID: PMC5484520
- DOI: 10.1371/journal.ppat.1006434
Genome-wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements
Abstract
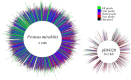
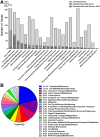
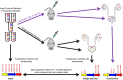
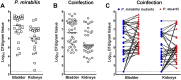
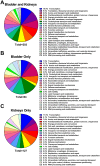
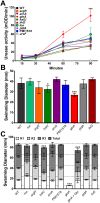
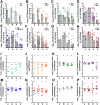
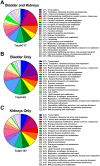
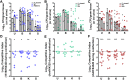
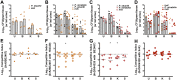
The Gram-negative bacterium Proteus mirabilis is a leading cause of catheter-associated urinary tract infections (CAUTIs), which are often polymicrobial. Numerous prior studies have uncovered virulence factors for P. mirabilis pathogenicity in a murine model of ascending UTI, but little is known concerning pathogenesis during CAUTI or polymicrobial infection. In this study, we utilized five pools of 10,000 transposon mutants each and transposon insertion-site sequencing (Tn-Seq) to identify the full arsenal of P. mirabilis HI4320 fitness factors for single-species versus polymicrobial CAUTI with Providencia stuartii BE2467. 436 genes in the input pools lacked transposon insertions and were therefore concluded to be essential for P. mirabilis growth in rich medium. 629 genes were identified as P. mirabilis fitness factors during single-species CAUTI. Tn-Seq from coinfection with P. stuartii revealed 217/629 (35%) of the same genes as identified by single-species Tn-Seq, and 1353 additional factors that specifically contribute to colonization during coinfection. Mutants were constructed in eight genes of interest to validate the initial screen: 7/8 (88%) mutants exhibited the expected phenotypes for single-species CAUTI, and 3/3 (100%) validated the expected phenotypes for polymicrobial CAUTI. This approach provided validation of numerous previously described P. mirabilis fitness determinants from an ascending model of UTI, the discovery of novel fitness determinants specifically for CAUTI, and a stringent assessment of how polymicrobial infection influences fitness requirements. For instance, we describe a requirement for branched-chain amino acid biosynthesis by P. mirabilis during coinfection due to high-affinity import of leucine by P. stuartii. Further investigation of genes and pathways that provide a competitive advantage during both single-species and polymicrobial CAUTI will likely provide robust targets for therapeutic intervention to reduce P. mirabilis CAUTI incidence and severity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Warren JW, Tenney JH, Hoopes JM, Muncie HL, Anthony WC. A Prospective Microbiologic Study of Bacteriuria in Patients with Chronic Indwelling Urethral Catheters. Journal of Infectious Diseases. 1982;146(6):719–23. doi: 10.1093/infdis/146.6.719 - DOI - PubMed
-
- Breitenbucher RB. Bacterial changes in the urine samples of patients with long-term indwelling catheters. Arch Intern Med. 1984;144(8):1585–8. Epub 1984/08/01. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

