Drugging the catalytically inactive state of RET kinase in RET-rearranged tumors
- PMID: 28615362
- PMCID: PMC5805089
- DOI: 10.1126/scitranslmed.aah6144
Drugging the catalytically inactive state of RET kinase in RET-rearranged tumors
Abstract
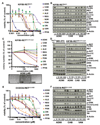
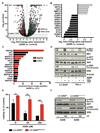
Oncogenic fusion events have been identified in a broad range of tumors. Among them, RET rearrangements represent distinct and potentially druggable targets that are recurrently found in lung adenocarcinomas. We provide further evidence that current anti-RET drugs may not be potent enough to induce durable responses in such tumors. We report that potent inhibitors, such as AD80 or ponatinib, that stably bind in the DFG-out conformation of RET may overcome these limitations and selectively kill RET-rearranged tumors. Using chemical genomics in conjunction with phosphoproteomic analyses in RET-rearranged cells, we identify the CCDC6-RETI788N mutation and drug-induced mitogen-activated protein kinase pathway reactivation as possible mechanisms by which tumors may escape the activity of RET inhibitors. Our data provide mechanistic insight into the druggability of RET kinase fusions that may be of help for the development of effective therapies targeting such tumors.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




References
-
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim Choi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. - PubMed
-
- Kodama T, Tsukaguchi T, Satoh Y, Yoshida M, Watanabe Y, Kondoh O, Sakamoto H. Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer. Molecular Cancer Therapeutics. 2014;13:2910–2918. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

