Comparative transcriptomes analysis of the wing disc between two silkworm strains with different size of wings
- PMID: 28617839
- PMCID: PMC5472328
- DOI: 10.1371/journal.pone.0179560
Comparative transcriptomes analysis of the wing disc between two silkworm strains with different size of wings
Abstract
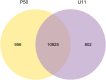
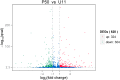
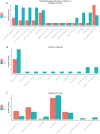
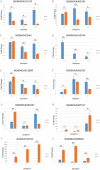
Wings of Bombyx mori (B. mori) develop from the primordium, and different B. mori strains have different wing types. In order to identify the key factors influencing B. mori wing development, we chose strains P50 and U11, which are typical for normal wing and minute wing phenotypes, respectively. We dissected the wing disc on the 1st-day of wandering stage (P50D1 and U11D1), 2nd-day of wandering stage (P50D2 and U11D2), and 3rd-day of wandering stage (P50D3 and U11D3). Subsequently, RNA-sequencing (RNA-Seq) was performed on both strains in order to construct their gene expression profiles. P50 exhibited 628 genes differentially expressed to U11, 324 up-regulated genes, and 304 down-regulated genes. Five enriched gene ontology (GO) terms were identified by GO enrichment analysis based on these differentially expressed genes (DEGs). KEGG enrichment analysis results showed that the DEGs were enriched in five pathways; of these, we identified three pathways related to the development of wings. The three pathways include amino sugar and nucleotide sugar metabolism pathway, proteasome signaling pathway, and the Hippo signaling pathway. The representative genes in the enrichment pathways were further verified by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The RNA-Seq and qRT-PCR results were largely consistent with each other. Our results also revealed that the significantly different genes obtained in our study might be involved in the development of the size of B. mori wings. In addition, several KEGG enriched pathways might be involved in the regulation of the pathways of wing formation. These results provide a basis for further research of wing development in B. mori.
Conflict of interest statement
Figures

References
-
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306(5703):1937–40. doi: 10.1126/science.1102210 . - DOI - PubMed
-
- Zhang YL, Xue RY, Cao GL, Zhu YX, Pan ZH, Gong CL. Shotgun proteomic analysis of wing discs from the domesticated silkworm (Bombyx mori) during metamorphosis. Amino acids. 2013;45(5):1231–41. doi: 10.1007/s00726-013-1588-8 . - DOI - PubMed
-
- Dubrovsky EB. Hormonal cross talk in insect development. Trends in endocrinology and metabolism: TEM. 2005;16(1):6–11. doi: 10.1016/j.tem.2004.11.003 . - DOI - PubMed
-
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Developmental cell. 2001;1(4):453–65. . - PubMed
-
- Ou J, Deng HM, Zheng SC, Huang LH, Feng QL, Liu L. Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC genomics. 2014;15:820 doi: 10.1186/1471-2164-15-820 ; - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

