Tag SNP selection for prediction of tick resistance in Brazilian Braford and Hereford cattle breeds using Bayesian methods
- PMID: 28619006
- PMCID: PMC5471684
- DOI: 10.1186/s12711-017-0325-2
Tag SNP selection for prediction of tick resistance in Brazilian Braford and Hereford cattle breeds using Bayesian methods
Abstract
Background: Cattle resistance to ticks is known to be under genetic control with a complex biological mechanism within and among breeds. Our aim was to identify genomic segments and tag single nucleotide polymorphisms (SNPs) associated with tick-resistance in Hereford and Braford cattle. The predictive performance of a very low-density tag SNP panel was estimated and compared with results obtained with a 50 K SNP dataset.
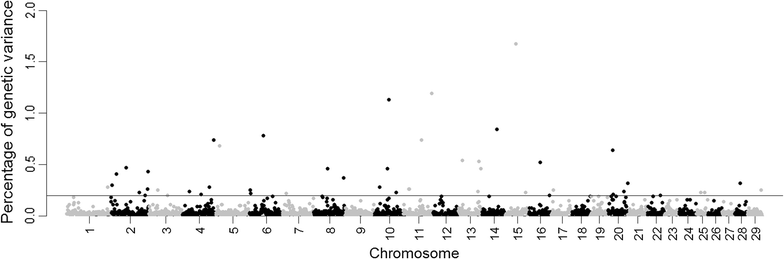
Results: BayesB (π = 0.99) was initially applied in a genome-wide association study (GWAS) for this complex trait by using deregressed estimated breeding values for tick counts and 41,045 SNP genotypes from 3455 animals raised in southern Brazil. To estimate the combined effect of a genomic region that is potentially associated with quantitative trait loci (QTL), 2519 non-overlapping 1-Mb windows that varied in SNP number were defined, with the top 48 windows including 914 SNPs and explaining more than 20% of the estimated genetic variance for tick resistance. Subsequently, the most informative SNPs were selected based on Bayesian parameters (model frequency and t-like statistics), linkage disequilibrium and minor allele frequency to propose a very low-density 58-SNP panel. Some of these tag SNPs mapped close to or within genes and pseudogenes that are functionally related to tick resistance. Prediction ability of this SNP panel was investigated by cross-validation using K-means and random clustering and a BayesA model to predict direct genomic values. Accuracies from these cross-validations were 0.27 ± 0.09 and 0.30 ± 0.09 for the K-means and random clustering groups, respectively, compared to respective values of 0.37 ± 0.08 and 0.43 ± 0.08 when using all 41,045 SNPs and BayesB with π = 0.99, or of 0.28 ± 0.07 and 0.40 ± 0.08 with π = 0.999.
Conclusions: Bayesian GWAS model parameters can be used to select tag SNPs for a very low-density panel, which will include SNPs that are potentially linked to functional genes. It can be useful for cost-effective genomic selection tools, when one or a few key complex traits are of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

