Molecular model of fission yeast centrosome assembly determined by superresolution imaging
- PMID: 28619713
- PMCID: PMC5551712
- DOI: 10.1083/jcb.201701041
Molecular model of fission yeast centrosome assembly determined by superresolution imaging
Abstract
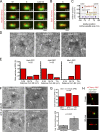
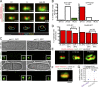
Microtubule-organizing centers (MTOCs), known as centrosomes in animals and spindle pole bodies (SPBs) in fungi, are important for the faithful distribution of chromosomes between daughter cells during mitosis as well as for other cellular functions. The cytoplasmic duplication cycle and regulation of the Schizosaccharomyces pombe SPB is analogous to centrosomes, making it an ideal model to study MTOC assembly. Here, we use superresolution structured illumination microscopy with single-particle averaging to localize 14 S. pombe SPB components and regulators, determining both the relationship of proteins to each other within the SPB and how each protein is assembled into a new structure during SPB duplication. These data enabled us to build the first comprehensive molecular model of the S. pombe SPB, resulting in structural and functional insights not ascertained through investigations of individual subunits, including functional similarities between Ppc89 and the budding yeast SPB scaffold Spc42, distribution of Sad1 to a ring-like structure and multiple modes of Mto1 recruitment.
© 2017 Bestul et al.
Figures





References
-
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
-
- Bouhlel I.B., Ohta M., Mayeux A., Bordes N., Dingli F., Boulanger J., Velve Casquillas G., Loew D., Tran P.T., Sato M., and Paoletti A.. 2015. Cell cycle control of spindle pole body duplication and splitting by Sfi1 and Cdc31 in fission yeast. J. Cell Sci. 128:1481–1493. 10.1242/jcs.159657 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

