Biological specificity of CDK4/6 inhibitors: dose response relationship, in vivo signaling, and composite response signature
- PMID: 28620137
- PMCID: PMC5546433
- DOI: 10.18632/oncotarget.18435
Biological specificity of CDK4/6 inhibitors: dose response relationship, in vivo signaling, and composite response signature
Abstract
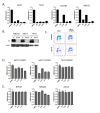
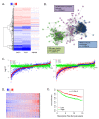
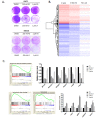
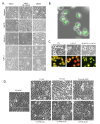
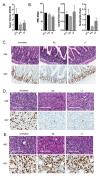
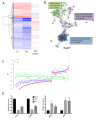
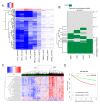
Recently developed potent and selective CDK4/6 inhibitors fall into two classes based on structure and toxicity profiles in clinical studies. One class, exemplified by palbociclib and ribociclib, exhibits neutropenia as a dose-limiting toxicity and requires discontinuous dosing. In contrast, the structurally distinct CDK4/6 inhibitor abemaciclib is dosed continuously, and has diarrhea and fatigue as dose-limiting toxicities. In preclinical models, palbociclib has been extensively studied and induces cell cycle inhibition in an RB-dependent manner. Thus far, abemaciclib has been less extensively evaluated. We found that abemaciclib cell cycle inhibitory activity is RB-dependent at clinically achievable concentrations. Abemaciclib elicited potent suppression of RB/E2F regulated genes associated with prognosis in ER-positive breast cancer. However, unlike palbociclib, at 250nM-1 µM doses abemaciclib induced cell death in RB-deficient cell lines. This response was associated with a rapidly-induced multi-vacuolar phenotype indicative of lysosomal membrane permeabilization that could be ameliorated with chloroquine. This event was not a reflection of inhibition of other CDK family members, but could be recapitulated with CBX4945 that inhibits casein and DYRK/HIPK kinases. To determine if these "off-target" features of abemaciclib were observed in vivo, mice harboring matched RB-positive and negative xenografts were treated with palbociclib and abemaciclib. In vivo, all of the apparent activity of abemaciclib was RB-dependent and strongly elicited suppression of cell cycle regulatory genes in a fashion markedly similar to palbociclib. Using gene expression data from cell lines and tumors treated with abemaciclib and palbociclib a composite signature of response to CDK4/6 inhibition was developed that included many genes that are individually required for tumor cell proliferation or viability. These data indicate that while abemaciclib and palbociclib can exert distinct biological and molecular effects, there are common gene expression features that could be broadly utilized in measuring the response to CDK4/6 inhibition.
Keywords: CDK4; E2F; abemaciclib; breast cancer; palbociclib.
Conflict of interest statement
This study was supported, in part, by sponsored research funding from Eli Lilly. The work was carried out independently in an academic setting and reflects the data and interpretations of the authors.
Figures
References
-
- Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6:353–67. doi: 10.1158/2159-8290.CD-15-0894. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

