Phenotypic, genotypic and antigenic characterization of emerging avian reoviruses isolated from clinical cases of arthritis in broilers in Saskatchewan, Canada
- PMID: 28620186
- PMCID: PMC5472580
- DOI: 10.1038/s41598-017-02743-8
Phenotypic, genotypic and antigenic characterization of emerging avian reoviruses isolated from clinical cases of arthritis in broilers in Saskatchewan, Canada
Abstract
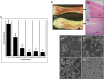
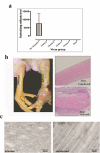
In recent years, emerging strains of pathogenic arthrogenic avian reovirus (ARV) have become a challenge to the chicken industry across USA and Canada causing significant economic impact. In this study, we characterized emerging variant ARV strains and examined their genetic and antigenic relationship with reference strains. We isolated 37 emerging variant ARV strains from tendons of broiler chickens with clinical cases of arthritis/tenosynovitis at commercial farms in Saskatchewan, Canada. Viral characterization using immunocytochemistry, gold-immunolabeling and electron microscopy revealed distinct features characteristic of ARV. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analyses of the viral Sigma C gene revealed genetic heterogeneity between the field isolates. On phylogenetic analyses, the Sigma C amino acid sequences of the isolates were clustered into four distinct genotypic groups. These ARV field strains were genetically diverse and quite distant from the vaccine and vaccine related field strains. Antibodies produced against a commercial Reo 2177 ® vaccine did not neutralize these variants. Moreover, structure based analysis of the Sigma C protein revealed significant antigenic variability between the cluster groups and the vaccine strains. To the best of our knowledge, this is the first report on the genetic, phenotypic and antigenic characterization of emerging ARVs in Canada.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Lee LH, Wang YH, Shien JH. Serological characterization of reoviruses isolated from avian species in Taiwan. J. Chin. Soc. Vet. Sci. 1992;18:69–72.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

