Molecular Detection of Tick-Borne Pathogen Diversities in Ticks from Livestock and Reptiles along the Shores and Adjacent Islands of Lake Victoria and Lake Baringo, Kenya
- PMID: 28620610
- PMCID: PMC5451513
- DOI: 10.3389/fvets.2017.00073
Molecular Detection of Tick-Borne Pathogen Diversities in Ticks from Livestock and Reptiles along the Shores and Adjacent Islands of Lake Victoria and Lake Baringo, Kenya
Abstract
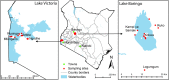
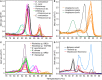
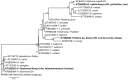
Although diverse tick-borne pathogens (TBPs) are endemic to East Africa, with recognized impact on human and livestock health, their diversity and specific interactions with tick and vertebrate host species remain poorly understood in the region. In particular, the role of reptiles in TBP epidemiology remains unknown, despite having been implicated with TBPs of livestock among exported tortoises and lizards. Understanding TBP ecologies, and the potential role of common reptiles, is critical for the development of targeted transmission control strategies for these neglected tropical disease agents. During the wet months (April-May; October-December) of 2012-2013, we surveyed TBP diversity among 4,126 ticks parasitizing livestock and reptiles at homesteads along the shores and islands of Lake Baringo and Lake Victoria in Kenya, regions endemic to diverse neglected tick-borne diseases. After morphological identification of 13 distinct Rhipicephalus, Amblyomma, and Hyalomma tick species, ticks were pooled (≤8 individuals) by species, host, sampling site, and collection date into 585 tick pools. By supplementing previously established molecular assays for TBP detection with high-resolution melting analysis of PCR products before sequencing, we identified high frequencies of potential disease agents of ehrlichiosis (12.48% Ehrlichia ruminantium, 9.06% Ehrlichia canis), anaplasmosis (6.32% Anaplasma ovis, 14.36% Anaplasma platys, and 3.08% Anaplasma bovis,), and rickettsiosis (6.15% Rickettsia africae, 2.22% Rickettsia aeschlimannii, 4.27% Rickettsia rhipicephali, and 4.95% Rickettsia spp.), as well as Paracoccus sp. and apicomplexan hemoparasites (0.51% Theileria sp., 2.56% Hepatozoon fitzsimonsi, and 1.37% Babesia caballi) among tick pools. Notably, we identified E. ruminantium in both Amblyomma and Rhipicephalus pools of ticks sampled from livestock in both study areas as well as in Amblyomma falsomarmoreum (66.7%) and Amblyomma nuttalli (100%) sampled from tortoises and Amblyomma sparsum (63.6%) sampled in both cattle and tortoises at Lake Baringo. Similarly, we identified E. canis in rhipicephaline ticks sampled from livestock and dogs in both regions and Amblyomma latum (75%) sampled from monitor lizards at Lake Victoria. These novel tick-host-pathogen interactions have implications on the risk of disease transmission to humans and domestic animals and highlight the complexity of TBP ecologies, which may include reptiles as reservoir species, in sub-Saharan Africa.
Keywords: Anaplasma; Babesia; Ehrlichia; Hepatozoon; Kenya; Rickettsia; Theileria; tick-borne diseases.
Figures


Similar articles
-
Pathogens, endosymbionts, and blood-meal sources of host-seeking ticks in the fast-changing Maasai Mara wildlife ecosystem.PLoS One. 2020 Aug 31;15(8):e0228366. doi: 10.1371/journal.pone.0228366. eCollection 2020. PLoS One. 2020. PMID: 32866142 Free PMC article.
-
Tissue-specific localization of tick-borne pathogens in ticks collected from camels in Kenya: insights into vector competence.Front Cell Infect Microbiol. 2024 Apr 18;14:1382228. doi: 10.3389/fcimb.2024.1382228. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38698904 Free PMC article.
-
Epidemiology of tick-borne pathogens in the semi-arid and the arid agro-ecological zones of Punjab province, Pakistan.Transbound Emerg Dis. 2019 Jan;66(1):526-536. doi: 10.1111/tbed.13059. Epub 2018 Nov 19. Transbound Emerg Dis. 2019. PMID: 30383917
-
Ticks and tick-borne pathogens of domestic animals in Somalia and neighbouring regions of Ethiopia and Kenya.Acta Trop. 2023 Jul;243:106944. doi: 10.1016/j.actatropica.2023.106944. Epub 2023 May 12. Acta Trop. 2023. PMID: 37178993 Review.
-
Hard ticks (Acari: Ixodidae) and tick-borne diseases of sheep and goats in Africa: A review.Ticks Tick Borne Dis. 2023 Nov;14(6):102232. doi: 10.1016/j.ttbdis.2023.102232. Epub 2023 Jul 31. Ticks Tick Borne Dis. 2023. PMID: 37531888 Review.
Cited by
-
Geographical distribution of ixodid ticks and tick-borne pathogens of domestic animals in Ethiopia: a systematic review.Parasit Vectors. 2022 Mar 28;15(1):108. doi: 10.1186/s13071-022-05221-x. Parasit Vectors. 2022. PMID: 35346354 Free PMC article.
-
Multi-country investigation of the diversity and associated microorganisms isolated from tick species from domestic animals, wildlife and vegetation in selected african countries.Exp Appl Acarol. 2021 Mar;83(3):427-448. doi: 10.1007/s10493-021-00598-3. Epub 2021 Mar 1. Exp Appl Acarol. 2021. PMID: 33646482 Free PMC article.
-
Molecular prevalence of emerging Anaplasma and Ehrlichia pathogens in apparently healthy dairy cattle in peri-urban Nairobi, Kenya.BMC Vet Res. 2020 Sep 29;16(1):364. doi: 10.1186/s12917-020-02584-0. BMC Vet Res. 2020. PMID: 32993638 Free PMC article.
-
Molecular Characterization of Rickettsial Agents in Ticks (Acari: Ixodidae) from Sri Lanka.Am J Trop Med Hyg. 2022 Apr 11;106(6):1613-23. doi: 10.4269/ajtmh.21-0995. Online ahead of print. Am J Trop Med Hyg. 2022. PMID: 35405644 Free PMC article.
-
Reptile vector-borne diseases of zoonotic concern.Int J Parasitol Parasites Wildl. 2021 Apr 22;15:132-142. doi: 10.1016/j.ijppaw.2021.04.007. eCollection 2021 Aug. Int J Parasitol Parasites Wildl. 2021. PMID: 34026483 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

