Structural insights into the function of ZRANB3 in replication stress response
- PMID: 28621305
- PMCID: PMC5481773
- DOI: 10.1038/ncomms15847
Structural insights into the function of ZRANB3 in replication stress response
Abstract
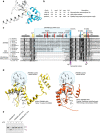
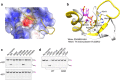
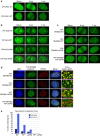
Strategies to resolve replication blocks are critical for the maintenance of genome stability. Among the factors implicated in the replication stress response is the ATP-dependent endonuclease ZRANB3. Here, we present the structure of the ZRANB3 HNH (His-Asn-His) endonuclease domain and provide a detailed analysis of its activity. We further define PCNA as a key regulator of ZRANB3 function, which recruits ZRANB3 to stalled replication forks and stimulates its endonuclease activity. Finally, we present the co-crystal structures of PCNA with two specific motifs in ZRANB3: the PIP box and the APIM motif. Our data provide important structural insights into the PCNA-APIM interaction, and reveal unexpected similarities between the PIP box and the APIM motif. We propose that PCNA and ATP-dependency serve as a multi-layered regulatory mechanism that modulates ZRANB3 activity at replication forks. Importantly, our findings allow us to interpret the functional significance of cancer associated ZRANB3 mutations.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Moldovan G. L., Pfander B. & Jentsch S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007). - PubMed
-
- Warbrick E. PCNA binding through a conserved motif. Bioessays 20, 195–199 (1998). - PubMed
-
- Bruning J. B. & Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure 12, 2209–2219 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

