CtIP/Ctp1/Sae2, molecular form fit for function
- PMID: 28623092
- PMCID: PMC5543718
- DOI: 10.1016/j.dnarep.2017.06.013
CtIP/Ctp1/Sae2, molecular form fit for function
Abstract
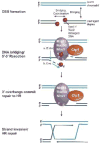
Vertebrate CtIP, and its fission yeast (Ctp1), budding yeast (Sae2) and plant (Com1) orthologs have emerged as key regulatory molecules in cellular responses to DNA double strand breaks (DSBs). By modulating the nucleolytic 5'-3' resection activity of the Mre11/Rad50/Nbs1 (MRN) DSB repair processing and signaling complex, CtIP/Ctp1/Sae2/Com1 is integral to the channeling of DNA double strand breaks through DSB repair by homologous recombination (HR). Nearly two decades since its discovery, emerging new data are defining the molecular underpinnings for CtIP DSB repair regulatory activities. CtIP homologs are largely intrinsically unstructured proteins comprised of expanded regions of low complexity sequence, rather than defined folded domains typical of DNA damage metabolizing enzymes and nucleases. A compact structurally conserved N-terminus forms a functionally critical tetrameric helical dimer of dimers (THDD) region that bridges CtIP oligomers, and is flexibly appended to a conserved C-terminal Sae2-homology DNA binding and DSB repair pathway choice regulatory hub which influences nucleolytic activities of the MRN core nuclease complex. The emerging evidence from structural, biophysical, and biological studies converges on CtIP having functional roles in DSB repair that include: 1) dynamic DNA strand coordination through direct DNA binding and DNA bridging activities, 2) MRN nuclease complex cofactor functions that direct MRN endonucleolytic cleavage of protein-blocked DSB ends and 3) acting as a protein binding hub targeted by the cell cycle regulatory apparatus, which influences CtIP expression and activity via layers of post-translational modifications, protein-protein interactions and DNA binding.
Keywords: CtIP/Ctp1/Sae2; DNA bridging; Homologous recombination; Intrinsically disordered proteins; Resection.
Published by Elsevier B.V.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

