Impact of Module-X2 and Carbohydrate Binding Module-3 on the catalytic activity of associated glycoside hydrolases towards plant biomass
- PMID: 28623337
- PMCID: PMC5473887
- DOI: 10.1038/s41598-017-03927-y
Impact of Module-X2 and Carbohydrate Binding Module-3 on the catalytic activity of associated glycoside hydrolases towards plant biomass
Abstract
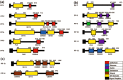
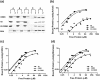
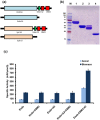
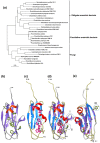
Cellulolytic enzymes capable of hydrolyzing plant biomass are secreted by microbial cells specifically in response to the carbon substrate present in the environment. These enzymes consist of a catalytic domain, generally appended to one or more non-catalytic Carbohydrate Binding Module (CBM), which enhances their activity towards recalcitrant biomass. In the present study, the genome of a cellulolytic microbe Paenibacillus polymyxa A18 was annotated for the presence of CBMs and analyzed their expression in response to the plant biomass and model polysaccharides Avicel, CMC and xylan using quantitative PCR. A gene that encodes X2-CBM3 was found to be maximally induced in response to the biomass and crystalline substrate Avicel. Association of X2-CBM3 with xyloglucanase and endoglucanase led to up to 4.6-fold increase in activity towards insoluble substrates. In the substrate binding study, module X2 showed a higher affinity towards biomass and phosphoric acid swollen cellulose, whereas CBM3 showed a higher affinity towards Avicel. Further structural modeling of X2 also indicated its potential role in substrate binding. Our findings highlighted the role of module X2 along with CBM3 in assisting the enzyme catalysis of agricultural residue and paved the way to engineer glycoside hydrolases for superior activity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

