Engineering of obligate intracellular bacteria: progress, challenges and paradigms
- PMID: 28626230
- PMCID: PMC5557331
- DOI: 10.1038/nrmicro.2017.59
Engineering of obligate intracellular bacteria: progress, challenges and paradigms
Abstract
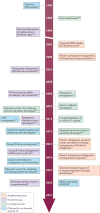
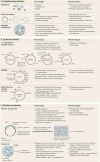
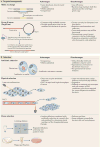
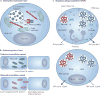
It is estimated that approximately one billion people are at risk of infection with obligate intracellular bacteria, but little is known about the underlying mechanisms that govern their life cycles. The difficulty in studying Chlamydia spp., Coxiella spp., Rickettsia spp., Anaplasma spp., Ehrlichia spp. and Orientia spp. is, in part, due to their genetic intractability. Recently, genetic tools have been developed; however, optimizing the genomic manipulation of obligate intracellular bacteria remains challenging. In this Review, we describe the progress in, as well as the constraints that hinder, the systematic development of a genetic toolbox for obligate intracellular bacteria. We highlight how the use of genetically manipulated pathogens has facilitated a better understanding of microbial pathogenesis and immunity, and how the engineering of obligate intracellular bacteria could enable the discovery of novel signalling circuits in host-pathogen interactions.
Figures
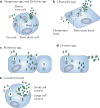

References
-
- Tam JE, Davis CH, Wyrick PB. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can. J. Microbiol. 1994;40:583–591. - PubMed
-
- Omsland A, Heinzen RA. Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu. Rev. Microbiol. 2011;65:111–128. This review describes the development of an axenic medium for C. burnetii, which enabled the rapid development of genetic tools for this microorganism. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI020384/AI/NIAID NIH HHS/United States
- R01 AI072683/AI/NIAID NIH HHS/United States
- U19 AI084044/AI/NIAID NIH HHS/United States
- R21 AI122014/AI/NIAID NIH HHS/United States
- T32 AI007540/AI/NIAID NIH HHS/United States
- R01 AI072606/AI/NIAID NIH HHS/United States
- R01 AI106859/AI/NIAID NIH HHS/United States
- R56 AI123346/AI/NIAID NIH HHS/United States
- R01 AI116523/AI/NIAID NIH HHS/United States
- R01 AI042792/AI/NIAID NIH HHS/United States
- R01 AI049424/AI/NIAID NIH HHS/United States
- R21 AI115449/AI/NIAID NIH HHS/United States
- R56 AI107951/AI/NIAID NIH HHS/United States
- R01 AI100759/AI/NIAID NIH HHS/United States
- R01 AI107951/AI/NIAID NIH HHS/United States
- R01 AI070908/AI/NIAID NIH HHS/United States
- R01 AI093653/AI/NIAID NIH HHS/United States
- R21 AI111086/AI/NIAID NIH HHS/United States
- R21 AI103272/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

