Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation
- PMID: 28627435
- PMCID: PMC5595636
- DOI: 10.1016/j.bbalip.2017.06.006
Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation
Abstract
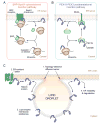
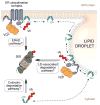
Lipid droplets (LDs) are ubiquitous, endoplasmic reticulum (ER)-derived organelles that mediate the sequestration of neutral lipids (e.g. triacylglycerol and sterol esters), providing a dynamic cellular storage depot for rapid lipid mobilization in response to increased cellular demands. LDs have a unique ultrastructure, consisting of a core of neutral lipids encircled by a phospholipid monolayer that is decorated with integral and peripheral proteins. The LD proteome contains numerous lipid metabolic enzymes, regulatory scaffold proteins, proteins involved in LD clustering and fusion, and other proteins of unknown functions. The cellular role of LDs is inherently determined by the composition of its proteome and alteration of the LD protein coat provides a powerful mechanism to adapt LDs to fluctuating metabolic states. Here, we review the current understanding of the molecular mechanisms that govern LD protein targeting and degradation. This article is part of a Special Issue entitled: Recent Advances in Lipid Droplet Biology edited by Rosalind Coleman and Matthijs Hesselink.
Keywords: Lipid droplet; Membrane protein; Protein degradation; Protein targeting; Proteome; Ubiquitin.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

