β-Catenin in desmoid-type fibromatosis: deep insights into the role of T41A and S45F mutations on protein structure and gene expression
- PMID: 28627792
- PMCID: PMC5664003
- DOI: 10.1002/1878-0261.12101
β-Catenin in desmoid-type fibromatosis: deep insights into the role of T41A and S45F mutations on protein structure and gene expression
Abstract
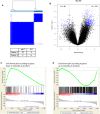
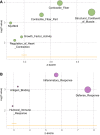
Desmoid-type fibromatosis (DF) is a rare mesenchymal lesion with high risk of local recurrence. Specific β-catenin mutations (S45F) appeared to be related to this higher risk compared to T41A-mutated or wild-type (WT). We explored the influence of both mutations and WT on structure stability and affinity of β-catenin for α-catenin and the pattern of gene expression that may influence DF behavior. Using 33 surgically resected primary DFs harboring T41A (n = 14), S45F (n = 10), or WT (n = 9), we performed a comparative molecular analysis by protein/protein interaction modeling, gene expression by DASL microarrays, human inflammation gene panel, and assessment of immune system-based biomarkers by immunohistochemistry. Mutated proteins were more stable than WT and formed a weaker complex with α-catenin. Consensus unsupervised gene clustering revealed the presence of two DF group-mutated (T41A + S45F) and WT (P = 0.0047). The gene sets 'Inflammatory-Defense-Humoral Immune Response' and 'Antigen Binding' were significantly enriched in T41A. The deregulation of 16 inflammation-related genes was confirmed. Low numbers of T cells and tumor-associated macrophages (TAM) infiltrating the tumors and low/absent PD-1/PD-L1 expression were also identified. We demonstrated that mutated DFs (T41A or S45F) and WT are two distinct molecular subgroups with regard to β-catenin stability, α-catenin affinity, and gene expression profiling. A different inflammation signature characterized the two mutated groups, suggesting mediation either by T41A or by S45F. Finally, all mutated cases showed a low number of TIL and TAM cells and a low or absent expression of PD-1 and PD-L1 consistent with β-catenin activation insensitive to checkpoint blockade.
Keywords: desmoid-type fibromatosis; gene expression; modeling; β-catenin mutation.
© 2017 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures


References
-
- Agresti A. (2007) Introduction to Categorical Data Analysis, 2nd edn. John Wiley & Sons, Inc., Hoboken, NJ.
-
- Bacac M, Migliavacca E, Stehle JC, McKee T, Delorenzi M, Coindre JM, Guillou L and Stamenkovic I (2006) A gene expression signature that distinguishes desmoid tumours from nodular fasciitis. J Pathol 208, 543–553. - PubMed
-
- Bonvalot S, Eldweny H, Haddad V, Rimareix F, Missenard G, Oberlin O, Vanel D, Terrier P, Blay JY, Le Cesne A et al (2008) Extra‐abdominal primary fibromatosis: aggressive management could be avoided in a subgroup of patients. Eur J Surg Oncol 34, 462–468. - PubMed
-
- Bozzi F, Conca E, Laurini E, Posocco P, Lo Sardo A, Jocollè G, Sanfilippo R, Gronchi A, Perrone F, Tamborini E et al (2013) In vitro and in silico studies of MDM2/MDMX isoforms predict Nutlin‐3A sensitivity in well/de‐differentiated liposarcomas. Lab Invest 93, 1232–1240. - PubMed
-
- van Broekhoven DL, Verhoef C, Grünhagen DJ, van Gorp JM, den Bakker MA, Hinrichs JW, de Voijs CM and van Dalen T (2015) Prognostic value of CTNNB1 gene mutation in primary sporadic aggressive fibromatosis. Ann Surg Oncol 22, 1464–1470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

