The ApaH-like phosphatase TbALPH1 is the major mRNA decapping enzyme of trypanosomes
- PMID: 28628654
- PMCID: PMC5491325
- DOI: 10.1371/journal.ppat.1006456
The ApaH-like phosphatase TbALPH1 is the major mRNA decapping enzyme of trypanosomes
Abstract
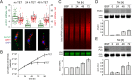
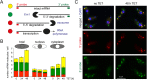
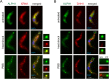
5'-3' decay is the major mRNA decay pathway in many eukaryotes, including trypanosomes. After deadenylation, mRNAs are decapped by the nudix hydrolase DCP2 of the decapping complex and finally degraded by the 5'-3' exoribonuclease. Uniquely, trypanosomes lack homologues to all subunits of the decapping complex, while deadenylation and 5'-3' degradation are conserved. Here, I show that the parasites use an ApaH-like phosphatase (ALPH1) as their major mRNA decapping enzyme. The protein was recently identified as a novel trypanosome stress granule protein and as involved in mRNA binding. A fraction of ALPH1 co-localises exclusively with the trypanosome 5'-3' exoribonuclease XRNA to a special granule at the posterior pole of the cell, indicating a connection between the two enzymes. RNAi depletion of ALPH1 is lethal and causes a massive increase in total mRNAs that are deadenylated, but have not yet started 5'-3' decay. These data suggest that ALPH1 acts downstream of deadenylation and upstream of mRNA degradation, consistent with a function in mRNA decapping. In vitro experiments show that recombinant, N-terminally truncated ALHP1 protein, but not a catalytically inactive mutant, sensitises the capped trypanosome spliced leader RNA to yeast Xrn1, but only if an RNA 5' polyphosphatase is included. This indicates that the decapping mechanism of ALPH1 differs from the decapping mechanism of Dcp2 by leaving more than one phosphate group at the mRNA's 5' end. This is the first reported function of a eukaryotic ApaH-like phosphatase, a bacterial-derived class of enzymes present in all phylogenetic super-groups of the eukaryotic kingdom. The substrates of eukaryotic ApaH-like phosphatases are unknown. However, the substrate of the related bacterial enzyme ApaH, diadenosine tetraphosphate, is highly reminiscent of a eukaryotic mRNA cap.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012. September;4(9):a012286 doi: 10.1101/cshperspect.a012286 - DOI - PMC - PubMed
-
- Kramer S. RNA in development: how ribonucleoprotein granules regulate the life cycles of pathogenic protozoa. WIREs RNA. 2014. March;5(2):263–84. doi: 10.1002/wrna.1207 - DOI - PubMed
-
- Lykke-Andersen J. Identification of a Human Decapping Complex Associated with hUpf Proteins in Nonsense-Mediated Decay. Mol Cell Biol. 2002. December 1;22(23):8114–21. doi: 10.1128/MCB.22.23.8114-8121.2002 - DOI - PMC - PubMed
-
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003. February;9(2):231–8. doi: 10.1261/rna.2151403 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

