Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases
- PMID: 28629429
- PMCID: PMC5477353
- DOI: 10.1186/s13073-017-0447-8
Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases
Abstract
Background: Genome rearrangements are critical oncogenic driver events in many malignancies. However, the identification and resolution of the structure of cancer genomic rearrangements remain challenging even with whole genome sequencing.
Methods: To identify oncogenic genomic rearrangements and resolve their structure, we analyzed linked read sequencing. This approach relies on a microfluidic droplet technology to produce libraries derived from single, high molecular weight DNA molecules, 50 kb in size or greater. After sequencing, the barcoded sequence reads provide long range genomic information, identify individual high molecular weight DNA molecules, determine the haplotype context of genetic variants that occur across contiguous megabase-length segments of the genome and delineate the structure of complex rearrangements. We applied linked read sequencing of whole genomes to the analysis of a set of synchronous metastatic diffuse gastric cancers that occurred in the same individual.
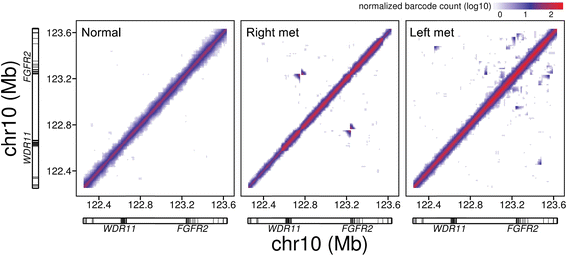
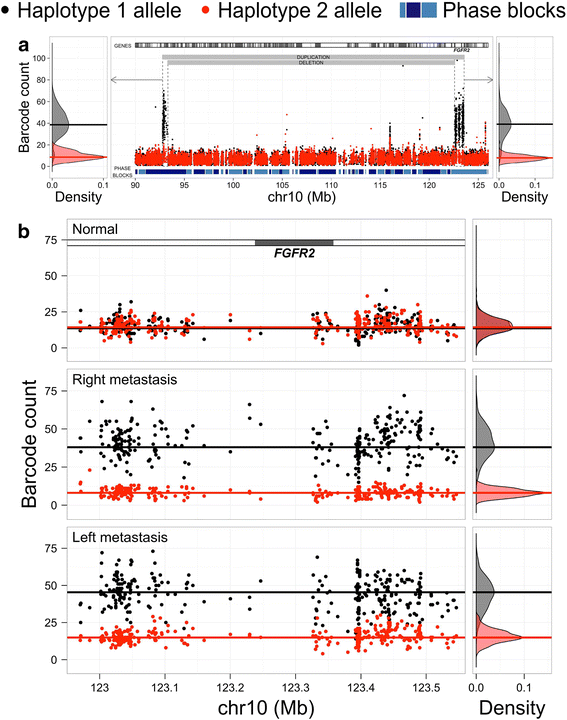
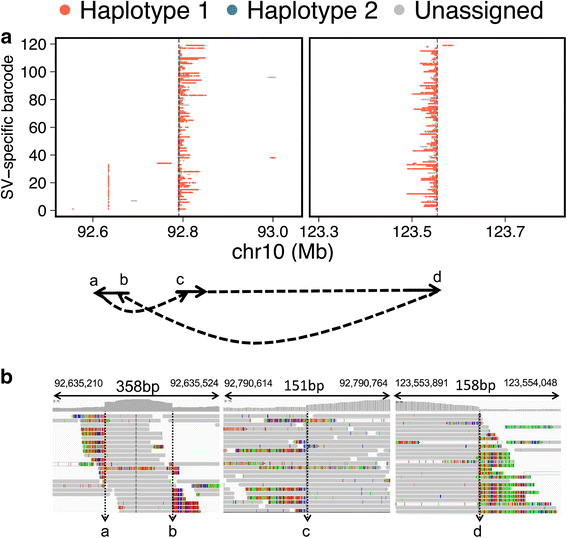
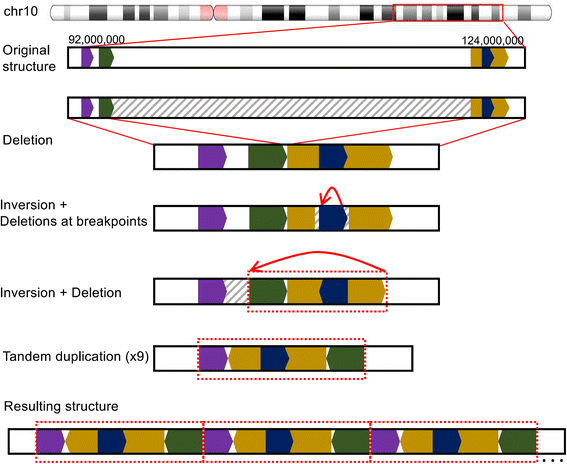
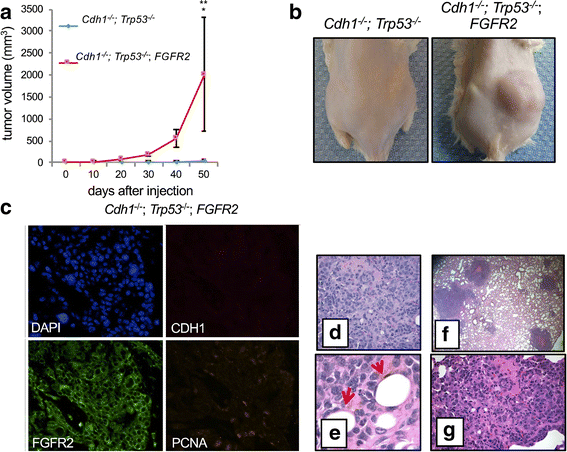
Results: When comparing metastatic sites, our analysis implicated a complex somatic rearrangement that was present in the metastatic tumor. The oncogenic event associated with the identified complex rearrangement resulted in an amplification of the known cancer driver gene FGFR2. With further investigation using these linked read data, the FGFR2 copy number alteration was determined to be a deletion-inversion motif that underwent tandem duplication, with unique breakpoints in each metastasis. Using a three-dimensional organoid tissue model, we functionally validated the metastatic potential of an FGFR2 amplification in gastric cancer.
Conclusions: Our study demonstrates that linked read sequencing is useful in characterizing oncogenic rearrangements in cancer metastasis.
Keywords: Barcode linked sequence reads; Cancer drivers; Cancer rearrangements; High molecular weight DNA; Whole genome analysis.
Figures
References
-
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

