Alzheimer's brains show inter-related changes in RNA and lipid metabolism
- PMID: 28630030
- PMCID: PMC5560656
- DOI: 10.1016/j.nbd.2017.06.008
Alzheimer's brains show inter-related changes in RNA and lipid metabolism
Abstract
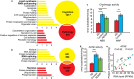
Alzheimer's disease (AD) involves changes in both lipid and RNA metabolism, but it remained unknown if these differences associate with AD's cognition and/or post-mortem neuropathology indices. Here, we report RNA-sequencing evidence of inter-related associations between lipid processing, cognition level, and AD neuropathology. In two unrelated cohorts, we identified pathway-enriched facilitation of lipid processing and alternative splicing genes, including the neuronal-enriched NOVA1 and hnRNPA1. Specifically, this association emerged in temporal lobe tissue samples from donors where postmortem evidence demonstrated AD neuropathology, but who presented normal cognition proximate to death. The observed changes further associated with modified ATP synthesis and mitochondrial transcripts, indicating metabolic relevance; accordingly, mass-spectrometry-derived lipidomic profiles distinguished between individuals with and without cognitive impairment prior to death. In spite of the limited group sizes, tissues from persons with both cognitive impairment and AD pathology showed elevation in several drug-targeted genes of other brain, vascular and autoimmune disorders, accompanied by pathology-related increases in distinct lipid processing transcripts, and in the RNA metabolism genes hnRNPH2, TARDBP, CLP1 and EWSR1. To further detect 3'-polyadenylation variants, we employed multiple cDNA primer pairs. This identified variants that showed limited differences in scope and length between the tested cohorts, yet enabled superior clustering of demented and non-demented AD brains versus controls compared to total mRNA expression values. Our findings indicate inter-related cognition-associated differences in AD's lipid processing, alternative splicing and 3'-polyadenylation, calling for pursuing the underlying psychological and therapeutics implications.
Keywords: Alternative polyadenylation; Alzheimer's disease; Cognitive decline; Lipidomics; Neuropathology; RNA sequencing.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

