Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states
- PMID: 28630140
- PMCID: PMC5558915
- DOI: 10.1261/rna.060632.117
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states
Abstract
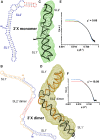
The 3'X domain is a 98-nt region located at the 3' end of hepatitis C virus genomic RNA that plays essential functions in the viral life cycle. It contains an absolutely conserved, 16-base palindromic sequence that promotes viral RNA dimerization, overlapped with a 7-nt tract implicated in a distal contact with a nearby functional sequence. Using small angle X-ray scattering measurements combined with model building guided by NMR spectroscopy, we have studied the stoichiometry, structure, and flexibility of domain 3'X and two smaller subdomain sequences as a function of ionic strength, and obtained a three-dimensional view of the full-length domain in its monomeric and dimeric states. In the monomeric form, the 3'X domain adopted an elongated conformation containing two SL1' and SL2' double-helical stems stabilized by coaxial stacking. This structure was significantly less flexible than that of isolated subdomain SL2' monomers. At higher ionic strength, the 3'X scattering envelope nearly doubled its size, reflecting the formation of extended homodimers containing an antiparallel SL2' duplex flanked by coaxially stacked SL1' helices. Formation of these dimers could initialize and/or regulate the packaging of viral RNA genomes into virions.
Keywords: 3′X domain; RNA; dimer; hepatitis C virus; small-angle X-ray scattering; structure.
© 2017 Cantero-Camacho et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
-
- Bartenschlager R, ed. 2013. Hepatitis C virus: from molecular virology to antiviral therapy. Springer-Verlag, Berlin.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
