Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells
- PMID: 28630143
- PMCID: PMC5496607
- DOI: 10.1083/jcb.201601109
Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells
Abstract
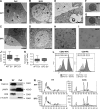
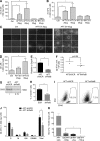
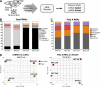
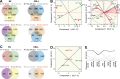
Extracellular vesicles (EVs) have been recently reported as crucial mediators in cell-to-cell communication in development and disease. In this study, we investigate whether mesenchymal stromal cells that constitute a supportive microenvironment for hematopoietic stem and progenitor cells (HSPCs) released EVs that could affect the gene expression and function of HSPCs. By taking advantage of two fetal liver-derived stromal lines with widely differing abilities to maintain HSPCs ex vivo, we demonstrate that stromal EVs play a critical role in the regulation of HSPCs. Both supportive and nonsupportive stromal lines secreted EVs, but only those delivered by the supportive line were taken up by HSPCs ex vivo and in vivo. These EVs harbored a specific molecular signature, modulated the gene expression in HSPCs after uptake, and maintained the survival and clonogenic potential of HSPCs, presumably by preventing apoptosis. In conclusion, our study reveals that EVs are an important component of the HSPC niche, which may have major applications in regenerative medicine.
© 2017 Stik et al.
Figures

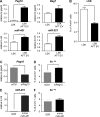
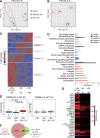
References
-
- Anzai N., Lee Y., Youn B.S., Fukuda S., Kim Y.J., Mantel C., Akashi M., and Broxmeyer H.E.. 2002. C-kit associated with the transmembrane 4 superfamily proteins constitutes a functionally distinct subunit in human hematopoietic progenitors. Blood. 99:4413–4421. 10.1182/blood.V99.12.4413 - DOI - PubMed
-
- Baglio S.R., Rooijers K., Koppers-Lalic D., Verweij F.J., Pérez Lanzón M., Zini N., Naaijkens B., Perut F., Niessen H.W., Baldini N., and Pegtel D.M.. 2015. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 6:127 10.1186/s13287-015-0116-z - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

