Topical Antimicrobial Treatments Can Elicit Shifts to Resident Skin Bacterial Communities and Reduce Colonization by Staphylococcus aureus Competitors
- PMID: 28630195
- PMCID: PMC5571303
- DOI: 10.1128/AAC.00774-17
Topical Antimicrobial Treatments Can Elicit Shifts to Resident Skin Bacterial Communities and Reduce Colonization by Staphylococcus aureus Competitors
Abstract
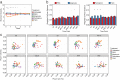
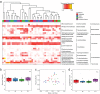
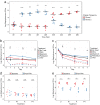
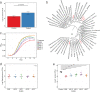
The skin microbiome is a complex ecosystem with important implications for cutaneous health and disease. Topical antibiotics and antiseptics are often employed to preserve the balance of this population and inhibit colonization by more pathogenic bacteria. However, despite their widespread use, the impact of these interventions on broader microbial communities remains poorly understood. Here, we report the longitudinal effects of topical antibiotics and antiseptics on skin bacterial communities and their role in Staphylococcus aureus colonization resistance. In response to antibiotics, cutaneous populations exhibited an immediate shift in bacterial residents, an effect that persisted for multiple days posttreatment. By contrast, antiseptics elicited only minor changes to skin bacterial populations, with few changes to the underlying microbiota. While variable in scope, both antibiotics and antiseptics were found to decrease colonization by commensal Staphylococcus spp. by sequencing- and culture-based methods, an effect which was highly dependent on baseline levels of Staphylococcus Because Staphylococcus residents have been shown to compete with the skin pathogen S. aureus, we also tested whether treatment could influence S. aureus levels at the skin surface. We found that treated mice were more susceptible to exogenous association with S. aureus and that precolonization with the same Staphylococcus residents that were previously disrupted by treatment reduced S. aureus levels by over 100-fold. In all, the results of this study indicate that antimicrobial drugs can alter skin bacterial residents and that these alterations can have critical implications for cutaneous host defense.
Keywords: Staphylococcus aureus; antimicrobial agents; microbiome; skin.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3:148–158. doi: 10.1038/mi.2009.132. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

