An Integrative Developmental Genomics and Systems Biology Approach to Identify an In Vivo Sox Trio-Mediated Gene Regulatory Network in Murine Embryos
- PMID: 28630873
- PMCID: PMC5467288
- DOI: 10.1155/2017/8932583
An Integrative Developmental Genomics and Systems Biology Approach to Identify an In Vivo Sox Trio-Mediated Gene Regulatory Network in Murine Embryos
Abstract
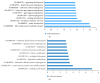
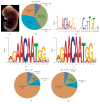
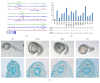
Embryogenesis is an intricate process involving multiple genes and pathways. Some of the key transcription factors controlling specific cell types are the Sox trio, namely, Sox5, Sox6, and Sox9, which play crucial roles in organogenesis working in a concerted manner. Much however still needs to be learned about their combinatorial roles during this process. A developmental genomics and systems biology approach offers to complement the reductionist methodology of current developmental biology and provide a more comprehensive and integrated view of the interrelationships of complex regulatory networks that occur during organogenesis. By combining cell type-specific transcriptome analysis and in vivo ChIP-Seq of the Sox trio using mouse embryos, we provide evidence for the direct control of Sox5 and Sox6 by the transcriptional trio in the murine model and by Morpholino knockdown in zebrafish and demonstrate the novel role of Tgfb2, Fbxl18, and Tle3 in formation of Sox5, Sox6, and Sox9 dependent tissues. Concurrently, a complete embryonic gene regulatory network has been generated, identifying a wide repertoire of genes involved and controlled by the Sox trio in the intricate process of normal embryogenesis.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

