ClinVar data parsing
- PMID: 28630944
- PMCID: PMC5473414
- DOI: 10.12688/wellcomeopenres.11640.1
ClinVar data parsing
Abstract
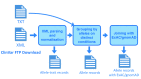
This software repository provides a pipeline for converting raw ClinVar data files into analysis-friendly tab-delimited tables, and also provides these tables for the most recent ClinVar release. Separate tables are generated for genome builds GRCh37 and GRCh38 as well as for mono-allelic variants and complex multi-allelic variants. Additionally, the tables are augmented with allele frequencies from the ExAC and gnomAD datasets as these are often consulted when analyzing ClinVar variants. Overall, this work provides ClinVar data in a format that is easier to work with and can be directly loaded into a variety of popular analysis tools such as R, python pandas, and SQL databases.
Keywords: ClinVar; Mendelian disease; XML parsing; pathogenic variants; variant interpretation.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources